Abstract
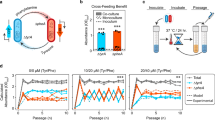
Cross-feeding interactions, in which bacterial cells exchange costly metabolites to the benefit of both interacting partners, are very common in the microbial world. However, it generally remains unclear what maintains this type of interaction in the presence of non-cooperating types. We investigate this problem using synthetic cross-feeding interactions: by simply deleting two metabolic genes from the genome of Escherichia coli, we generated genotypes that require amino acids to grow and release other amino acids into the environment. Surprisingly, in a vast majority of cases, cocultures of two cross-feeding strains showed an increased Darwinian fitness (that is, rate of growth) relative to prototrophic wild type cells—even in direct competition. This unexpected growth advantage was due to a division of metabolic labour: the fitness cost of overproducing amino acids was less than the benefit of not having to produce others when they were provided by their partner. Moreover, frequency-dependent selection maintained cross-feeding consortia and limited exploitation by non-cooperating competitors. Together, our synthetic study approach reveals ecological principles that can help explain the widespread occurrence of obligate metabolic cross-feeding interactions in nature.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Amarasekare P . (2003). Competitive coexistence in spatially structured environments: a synthesis. Ecol Lett 6: 1109–1122.
Axelrod R, Hamilton WD . (1981). The evolution of cooperation. Science 211: 1390–1396.
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 0008.
Benjamini Y, Hochberg Y . (2000). On the adaptive control of the false discovery fate in multiple testing with independent statistics. J Educ Behav Stat 25: 60–83.
Bertels F, Merker H, Kost C . (2012). Design and characterization of auxotrophy-based amino acid biosensors. PLoS One 7: e41349.
Bohl K, de Figueiredo LF, Hädicke O, Klamt S, Kost C, Schuster S, Kaleta C . (2010). CASOP GS: Computing intervention strategies targeted at production improvement in genome-scale metabolic networks. . In: Schomburg D, Grote A (eds). Proceedings of the 25th German Conference on Bioinformatics. Gesellschaft f. Informatik: Bonn, 2010, pp 71–80 (ISSN: 1617-5468).
Boucher DH . (1988) The Biology of Mutualism: Ecology and Evolution. Oxford University Press: New York.
Bull JJ, Rice WR . (1991). Distinguishing mechanisms for the evolution of cooperation. J Theor Biol 149: 63–74.
Damore JA, Gore J . (2012). Understanding microbial cooperation. J Theor Biol 299: 31–41.
Datsenko K, Wanner B . (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645.
Doebeli M . (2002). A model for the evolutionary dynamics of cross-feeding polymorphisms in microorganisms. Popul Ecol 44: 59–70.
Doebeli M, Knowlton N . (1998). The evolution of interspecific mutualisms. Proc Natl Acad Sci USA 95: 8676–8680.
Douglas AE . (1994) Symbiotic Interactions. Oxford University Press: New York.
Ferriere R, Bronstein JL, Rinaldi S, Law R, Gauduchon M . (2002). Cheating and the evolutionary stability of mutualisms. Proc R Soc Lond B Biol Sci 269: 773–780.
Ferriere R, Gauduchon M, Bronstein JL . (2007). Evolution and persistence of obligate mutualists and exploiters: competition for partners and evolutionary immunization. Ecol Lett 10: 115–126.
Foster KR, Bell T . (2012). Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol 22: 1845–1850.
Friesen ML, Saxer G, Travisano M, Doebeli M . (2004). Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58: 245–260.
Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M et al. (2001). Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291: 2606–2608.
Harcombe W . (2010). Novel cooperation experimentally evolved between species. Evolution 64: 2166–2172.
Hillesland KL, Stahl DA . (2010). Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci USA 107: 2124–2129.
Hosoda K, Suzuki S, Yamauchi Y, Shiroguchi Y, Kashiwagi A, Ono N et al. (2011). Cooperative adaptation to establishment of a synthetic bacterial mutualism. PLoS One 6: e17105.
Hottes AK, Freddolino PL, Khare A, Donnell ZN, Liu JC, Tavazoie S . (2013). Bacterial adaptation through loss of function. PLoS Genet 9: e1003617.
Huynen M, Dandekar T, Bork P . (1999). Variation and evolution of the citric acid cycle: a genomic perspective. Trends Microbiol 7: 281–291.
Ishii S, Kosaka T, Hori K, Hotta Y, Watanabe K . (2005). Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus. Appl Environ Microbiol 71: 7838–7845.
Koskiniemi S, Sun S, Berg OG, Andersson DI . (2012). Selection-driven gene loss in bacteria. PLoS Genet 8: e1002787.
Lee MC, Marx CJ . (2012). Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet 8: e1002651.
Lenski RE, Rose MR, Simpson SC, Tadler SC . (1991). Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138: 1315–1341.
Ley RE, Harris JK, Wilcox J, Spear JR, Miller SR, Bebout BM et al. (2006). Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl Environ Microbiol 72: 3685–3695.
Little AEF, Robinson CJ, Peterson SB, Raffa KE, Handelsman J . (2008). Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol 62: 375–401.
Maharjan RP, Ferenci T . (2005). Metabolomic diversity in the species Escherichia coli and its relationship to genetic population structure. Metabolomics 1: 235–242.
Maharjan RP, Ferenci T, Reeves PR, Li Y, Liu B, Wang L . (2012). The multiplicity of divergence mechanisms in a single evolving population. Genome Biol 13: r41.
McCutcheon JP, Moran NA . (2012). Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10: 13–26.
McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L et al. (2007). The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci USA 104: 7600–7605.
McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJM, Schink B et al. (2008). Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann NY Acad Sci 1125: 58–72.
Morris BE, Henneberger R, Huber H, Moissl-Eichinger C . (2013). Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev 37: 384–406.
Nauhaus K, Boetius A, Kruger M, Widdel F . (2002). In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ Microbiol 4: 296–305.
Orphan VJ . (2009). Methods for unveiling cryptic microbial partnerships in nature. Curr Opin Microbiol 12: 231–237.
Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ . (2008). Diverse syntrophic partnerships from-deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci USA 105: 7052–7057.
Phelan VV, Liu W-T, Pogliano K, Dorrestein PC . (2012). Microbial metabolic exchange—the chemotype-to-phenotype link. Nat Chem Biol 8: 26–35.
Poltak S, Cooper V . (2011). Ecological succession in long-term experimentally evolved biofilms produces synergistic communities. ISME J 5: 369–378.
Rosenzweig RF, Sharp RR, Treves DS, Adams J . (1994). Microbial evolution in a simple unstructured environment:genetic differentiation in Escherichia coli. Genetics 137: 903–917.
Rueffler C, Hermisson J, Wagner GP . (2012). Evolution of functional specialization and division of labor. Proc Natl Acad Sci USA 109: E326–E335.
Sachs JL, Mueller UG, Wilcox TP, Bull JJ . (2004). The evolution of cooperation. Q Rev Biol 79: 135–160.
Sachs JL, Simms EL . (2006). Pathways to mutualism breakdown. Trends Ecol Evol 21: 585–592.
Schink B . (2002). Synergistic interactions in the microbial world. Antonie Leeuwenhoek 81: 257–261.
Sieuwerts S, FAM de Bok, Hugenholtz J, van Hylckama Vlieg JET . (2008). Unraveling microbial interactions in food fermentations; from classical to genomics approaches. Appl Environ Microbiol 74: 4997–5007.
Thomason L, Costantino N, Court DL . (2007). E. coli genome manipulation by P1 transduction. Curr Prot Mol Biol 1: 17.
Vanstockem M, Michiels K, Vanderleyden J, Vangool A . (1987). Transposon mutagenesis of Azospirillum brasilense and Azospirillum lipoferum:physical analysis of Tn5 and Tn5-Mob insertion mutants. Appl Environ Microb 53: 410–415.
Vellend M . (2010). Conceptual synthesis in community ecology. Q Rev Biol 85: 183–206.
Wahl LM . (2002). Evolving the division of labour: generalists, specialists and task allocation. J Theor Biol 219: 371–388.
Wanner G, Vogl K, Overmann J . (2008). Ultrastructural characterization of the prokaryotic symbiosis in ‘Chlorochromatium aggregatum’. J Bacteriol 190: 3721–3730.
Wilkinson TG, Topiwala HH, Hamer G . (1974). Interactions in a mixed bacterial population growing on methane in continuous culture. Biotechnol Bioeng 16: 41–59.
Wilson WG, Morris WF, Bronstein JL . (2003). Coexistence of mutualists and exploiters on spatial landscapes. Ecol Monogr 73: 397–413.
Wintermute E, Silver P . (2010). Emergent cooperation in microbial metabolism. Mol Syst Biol 6: 407.
Yu DW . (2001). Parasites of mutualisms. Biol J Linnean Soc 72: 529–546.
Zhang Y, Laing C, Steele M, Ziebell K, Johnson R, Benson A et al. (2007). Genome evolution in major Escherichia coli O157: H7 lineages. BMC Genomics 8: 121.
Acknowledgements
We would like to thank W Boland and J Gershenzon for support and IT Baldwin, TF Cooper, K Hammerschmidt, M Kaltenpoth, E Kothe, E Libby, PB Rainey, W Ratcliff, S Schmidt, and two anonymous reviewers for helpful discussions and constructive criticism of the manuscript. This work was supported by the Jena School for Microbial Communication (C Kaleta, C Kost, SP, HM and SS), International Max Planck Research School (KB, C Kaleta, C Kost and SS), Volkswagen Foundation (C Kost), Max Planck Society (C Kost, MR), Fundacão Calouste Gulbenkian (LFdF), Fundacão para a Ciencia e a Tecnologia (LFdF), and Siemens SA Portugal (LFdF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The Authors declare no conflict of interest.
Additional information
Conceived the project: SP and C Kost. Designed the experiments: SP, C Kaleta and C Kost. Performed all experiments: SP. Developed the AA measurement protocol: HM and MR. Independently replicated some experiments (data not shown): HM. Performed AA measurements: HM, SP, and MR. Analysed the data: SP and C Kost. Developed and applied the CASOP-GS method: C Kaleta, KB, LFdF, and SS. Wrote the paper: SP and C Kost. Amended the manuscript: all authors.
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Pande, S., Merker, H., Bohl, K. et al. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J 8, 953–962 (2014). https://doi.org/10.1038/ismej.2013.211
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2013.211
Keywords
This article is cited by
-
CoNoS: synthetische Ko-Kulturen für Grundlagenforschung und Anwendung
BIOspektrum (2024)
-
Metabolic exchanges are ubiquitous in natural microbial communities
Nature Microbiology (2023)
-
Bacteriophage-mediated lysis supports robust growth of amino acid auxotrophs
The ISME Journal (2023)
-
Stress-induced metabolic exchanges between complementary bacterial types underly a dynamic mechanism of inter-species stress resistance
Nature Communications (2023)
-
Impact of direct physical association and motility on fitness of a synthetic interkingdom microbial community
The ISME Journal (2023)