Abstract
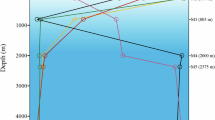
Bacterioplankton of the SAR11 clade are the most abundant microorganisms in marine systems, usually representing 25% or more of the total bacterial cells in seawater worldwide. SAR11 is divided into subclades with distinct spatiotemporal distributions (ecotypes), some of which appear to be specific to deep water. Here we examine the genomic basis for deep ocean distribution of one SAR11 bathytype (depth-specific ecotype), subclade Ic. Four single-cell Ic genomes, with estimated completeness of 55%–86%, were isolated from 770 m at station ALOHA and compared with eight SAR11 surface genomes and metagenomic datasets. Subclade Ic genomes dominated metagenomic fragment recruitment below the euphotic zone. They had similar COG distributions, high local synteny and shared a large number (69%) of orthologous clusters with SAR11 surface genomes, yet were distinct at the 16S rRNA gene and amino-acid level, and formed a separate, monophyletic group in phylogenetic trees. Subclade Ic genomes were enriched in genes associated with membrane/cell wall/envelope biosynthesis and showed evidence of unique phage defenses. The majority of subclade Ic-specfic genes were hypothetical, and some were highly abundant in deep ocean metagenomic data, potentially masking mechanisms for niche differentiation. However, the evidence suggests these organisms have a similar metabolism to their surface counterparts, and that subclade Ic adaptations to the deep ocean do not involve large variations in gene content, but rather more subtle differences previously observed deep ocean genomic data, like preferential amino-acid substitutions, larger coding regions among SAR11 clade orthologs, larger intergenic regions and larger estimated average genome size.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Abascal F, Zardoya R, Posada D . (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105.
Aguey-Zinsou K-F, Bernhardt PV, Kappler U, McEwan AG . (2003). Direct electrochemistry of a bacterial sulfite dehydrogenase. J Am Chem Soc 125: 530–535.
Anders S, Huber W . (2010). Differential expression analysis for sequence count data. Genome Biol 11: R106.
Arıstegui J, Gasol JM, Duarte CM, Herndl GJ . (2009). Microbial oceanography of the dark ocean’s pelagic realm. Limnol Oceanogr 54: 1501–1529.
Blainey PC . (2013). The future is now: single-cell genomics of bacteria and archaea. FEMS Microbiol Rev 37: 407–427.
Brown MV, Lauro FM, DeMaere MZ, Les M, Wilkins D, Thomas T et al (2012). Global biogeography of SAR11 marine bacteria. Mol Sys Biol 8: 1–13.
Campanaro S, Treu L, Valle G . (2008). Protein evolution in deep sea bacteria: an analysis of amino acids substitution rates. BMC Evol Biol 8: 313.
Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, DeLong EF et al (2010). A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science 330: 1375–1378.
Carini P, Steindler L, Beszteri S, Giovannoni SJ . (2012). Nutrient requirements for growth of the extreme oligotroph 'Candidatus Pelagibacter ubique' HTCC1062 on a defined medium. ISME J 7: 592–602.
Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K . (2009). Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3: 283–295.
Castresana J . (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552.
Chou HH, Holmes MH . (2001). DNA sequence quality trimming and vector removal. Bioinformatics 17: 1093–1104.
Coleman ML . (2006). Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770.
DeLong EF, Yayanos AA . (1985). Adaptation of the membrane lipids of a deep-sea bacterium to changes in hydrostatic pressure. Science 228: 1101–1103.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496.
Eddy SR . (2011). Accelerated Profile HMM Searches. PLoS Comput Biol 7: e1002195.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Eloe EA, Fadrosh DW, Novotny M, Zeigler Allen L, Kim M, Lombardo M-J et al (2011a). Going deeper: metagenome of a hadopelagic microbial community. PLoS One 6: e20388.
Eloe EA, Shulse CN, Fadrosh DW, Williamson SJ, Allen EE, Bartlett DH . (2011b). Compositional differences in particle‐associated and free‐living microbial assemblages from an extreme deep‐ocean environment. Environ Microbiol Rep 3: 449–458.
Field K, Gordon D, Wright T, Rappe M, Urbach E, Vergin K et al (1997). Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol 63: 63–70.
Fu L, Niu B, Zhu Z, Wu S, Li W . (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152.
Giovannoni SJ, Rappe MS, Vergin KL, Adair NL . (1996). 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA 93: 7979–7984.
Giovannoni SJ, Vergin KL . (2012). Seasonality in ocean microbial communities. Science 335: 671–676.
Gordon DA, Giovannoni SJ . (1996). Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol 62: 1171–1177.
Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ et al (2012). Streamlining and core genome conservation among highly divergent members of the SAR11 Clade. mBio 3: e00252–00212.
Haft DH, Selengut J, Mongodin EF, Nelson KE . (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLOS Comput Biol 1: e60.
Hay NA, Tipper DJ, Gygi D, Hughes C . (1999). A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol 181: 2008–2016.
Ivars-Martínez E, Martin-Cuadrado A-B, D'Auria G, Mira A, Ferriera S, Johnson J et al (2008). Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J 2: 1194–1212.
Kappler U, Bennett B, Rethmeier J, Schwarz G, Deutzmann R, McEwan AG et al (2000). Sulfite: cytochrome c oxidoreductase from Thiobacillus novellus. J Biol Chem 275: 13202–13212.
Kappler U, Davenport K, Beatson S, Lucas S, Lapidus A, Copeland A et al (2012). Complete genome sequence of the facultatively chemolithoautotrophic and methylotrophic alpha Proteobacterium Starkeya novella type strain (ATCC 8093(T)). Stand Genomic Sci 7: 44–58.
Karner MB, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510.
King GM, Smith CB, Tolar B, Hollibaugh JT . (2013). Analysis of composition and structure of coastal to mesopelagic bacterioplankton communities in the northern gulf of Mexico. Front Microbiol 3: 438.
Konstantinidis KT, Tiedje JM . (2007). Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol 10: 504–509.
Konstantinidis KT, Braff J, Karl DM, DeLong EF . (2009). Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol 75: 5345–5355.
Lasken RS . (2013). Single-cell sequencing in its prime. Nat Biotechnol 31: 211–212.
Lauro FM, Bartlett DH . (2008). Prokaryotic lifestyles in deep sea habitats. Extremophiles 12: 15–25.
Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P et al (2011). Evolution and classification of the CRISPR/Cas systems. Nat Rev Micro 9: 467–477.
Martin-Cuadrado A-B, López-García P, Alba J-C, Moreira D, Monticelli L, Strittmatter A et al (2007). Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS One 2: e914.
Madigan MT, Martinko JM . (2006) Brock Biology of Microorganisms, 11th edn. Pearson Prentice Hall: Upper Saddle River, NJ, USA.
Morris R, Rappé M, Connon S, Vergin K, Siebold WA, Carlson CA et al (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810.
Morris RM, Longnecker K, Giovannoni SJ . (2006). Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol 8: 1361–1370.
Morris RM, Frazar CD, Carlson CA . (2012). Basin-scale patterns in the abundance of SAR11 subclades, marine Actinobacteria (OM1), members of the Roseobacter clade and OCS116 in the South Atlantic. Environ Microbiol 14: 1133–1144.
Nagata T, Tamburini C, Arístegui J, Baltar F, Bochdansky AB, Fonda-Umani S et al (2010). Emerging concepts on microbial processes in the bathypelagic ocean – ecology, biogeochemistry, and genomics. Deep-Sea Res II 57: 1519–1536.
Pester M, Schleper C, Wagner M . (2011). The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14: 300–306.
Quaiser A, Zivanovic Y, Moreira D, López-García P . (2010). Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5: 285–304.
Reinthaler T, van Aken HM, Herndl GJ . (2010). Major contribution of autotrophy to microbial carbon cycling in the deep North Atlantic’s interior. Deep-Sea Res II 57: 1572–1580.
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F et al (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437.
Robbertse B, Yoder RJ, Boyd A, Reeves J, Spatafora JW . (2011). Hal: an automated pipeline for phylogenetic analyses of genomic data. PLoS Curr Tree Life 3: RRN1213.
Robinson C, Steinberg DK, Anderson TR, Arístegui J, Carlson CA, Frost JR et al (2010). Mesopelagic zone ecology and biogeochemistry–a synthesis. Deep-Sea Res II 57: 1504–1518.
Rodriguez-Valera F, Martin-Cuadrado A-B, Rodriguez-Brito B, Pašić L, Thingstad TF, Rohwer F et al (2009). Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7: 828–836.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al (2007). The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5: e77.
Schattenhofer M, Fuchs BM, Amann R, Zubkov MV, Tarran GA, Pernthaler J . (2009). Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ Microbiol 11: 2078–2093.
Schwalbach MS, Tripp HJ, Steindler L, Smith DP, Giovannoni SJ . (2010). The presence of the glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ Microbiol 12: 490–500.
Shi Y, Tyson GW, Eppley JM, DeLong EF . (2011). Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5: 999–1013.
Simonato F, Campanaro S, Lauro FM, Vezzi A, D'Angelo M, Vitulo N et al (2006). Piezophilic adaptation: a genomic point of view. J Biotechnol 126: 11–25.
Smedile F, Messina E, La Cono V, Tsoy O, Monticelli LS, Borghini M et al (2013). Metagenomic analysis of hadopelagic microbial assemblages thriving at the deepest part of Mediterranean Sea, Matapan-Vavilov Deep. Environ Microbiol 15: 167–182.
Stamatakis A . (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Stamatakis A, Hoover P, Rougemont J . (2008). A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol 57: 758–771.
Stepanauskas R . (2012). Single cell genomics: an individual look at microbes. Curr Opin Microbiol 15: 613–620.
Stewart FJ, Ulloa O, DeLong EF . (2012). Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol 14: 23–40.
Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the Dark Ocean. Science 333: 1296–1300.
Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ et al (2011). Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep 1: 1–9.
Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA et al (2009). Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J 3: 1148–1163.
Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ . (2008). SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452: 741–744.
Tully BJ, Nelson WC, Heidelberg JF . (2011). Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine. Environ Microbiol 14: 254–267.
Varela MM, Van Aken HM, Herndl GJ . (2008). Abundance and activity of Chloroflexi‐type SAR202 bacterioplankton in the meso- and bathypelagic waters of the (sub) tropical Atlantic. Environ Microbiol 10: 1903–1911.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Vergin KL, Tripp HJ, Wilhelm LJ, Denver DR, Rappé MS, Giovannoni SJ . (2007). High intraspecific recombination rate in a native population of Candidatus Pelagibacter ubique (SAR11). Environ Microbiol 9: 2430–2440.
Vergin KL, Beszteri B, Monier A, Thrash JC, Temperton B, Treusch AH et al (2013). High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series Study site by phylogenetic placement of pyrosequences. ISME J 7: 1322–1332.
Viklund J, Ettema TJG, Andersson SGE . (2012). Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol Biol Evol 29: 599–615.
Vos M, Didelot X . (2009). A comparison of homologous recombination rates in bacteria and archaea. ISME J 3: 199–208.
Wang F, Wang J, Jian H, Zhang B, Li S, Wang F et al (2008). Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS One 3: e1937.
Westesson O, Barquist L, Holmes I . (2012). HandAlign: Bayesian multiple sequence alignment, phylogeny and ancestral reconstruction. Bioinformatics 28: 1170–1171.
Wilhelm LJ, Tripp HJ, Givan SA, Smith DP, Giovannoni SJ . (2007). Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol Direct 2: 27.
Williams TJ, Long E, Evans F, DeMaere MZ, Lauro FM, Raftery MJ et al (2012). A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J 6: 1883–1900.
Wright TD, Vergin KL, Boyd PW, Giovannoni SJ . (1997). A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol 63: 1441–1448.
Yelton AP, Thomas BC, Simmons SL, Wilmes P, Zemla A, Thelen MP et al (2011). A semi-quantitative, synteny-based method to improve functional predictions for hypothetical and poorly annotated bacterial and archaeal genes. PLoS Comput Biol 7: e1002230.
Yilmaz P, Gilbert JA, Knight R, Amaral-Zettler L, Karsch-Mizrachi I, Cochrane G et al (2011). The genomic standards consortium: bringing standards to life for microbial ecology. ISME J 5: 1565–1567.
Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC et al (2013). Abundant SAR11 viruses in the ocean. Nature 494: 357–360.
Acknowledgements
This work was supported by the Gordon and Betty Moore Foundation (SJG and EFD), the US Department of Energy Joint Genome Institute (JGI) Community Supported Program grant 2011-387 (RS, BKS, EFD, SJG), National Science Foundation (NSF) Science and Technology Center Award EF0424599 (EFD), NSF awards EF-826924 (RS), OCE-821374 (RS) and OCE-1232982 (RS and BKS), and is based on work supported by the NSF under Award no. DBI-1003269 (JCT). Sequencing was conducted by JGI and supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH11231. We thank Christopher M Sullivan and the Oregon State University Center for Genome Research and Biocomputing, as well as the Louisiana State University Center for Computation and Technology for vital computational resources. We also thank Kelly C Wrighton and Laura A Hug for critical discussions about single-cell genomics, metagenomics and metabolic reconstruction.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Cameron Thrash, J., Temperton, B., Swan, B. et al. Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype. ISME J 8, 1440–1451 (2014). https://doi.org/10.1038/ismej.2013.243
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2013.243
Keywords
This article is cited by
-
Ecophysiology and genomics of the brackish water adapted SAR11 subclade IIIa
The ISME Journal (2023)
-
Genome-based characterization of the deep-sea psychrotolerant bacterium Bacillus altitudinis SORB11 isolated from the Indian Sector of the Southern Ocean
Polar Biology (2023)
-
Microdiversity characterizes prevalent phylogenetic clades in the glacier-fed stream microbiome
The ISME Journal (2022)
-
Distinctive gene and protein characteristics of extremely piezophilic Colwellia
BMC Genomics (2020)
-
Diversity and biogeography of SAR11 bacteria from the Arctic Ocean
The ISME Journal (2020)