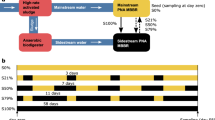
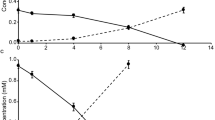
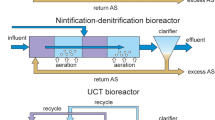
Abstract
Numerous past studies have shown members of the genus Nitrospira to be the predominant nitrite-oxidizing bacteria (NOB) in nitrifying wastewater treatment plants (WWTPs). Only recently, the novel NOB ‘Candidatus Nitrotoga arctica’ was identified in permafrost soil and a close relative was enriched from activated sludge. Still, little is known about diversity, distribution and functional importance of Nitrotoga in natural and engineered ecosystems. Here we developed Nitrotoga 16S rRNA-specific PCR primers and fluorescence in situ hybridization (FISH) probes, which were applied to screen activated sludge samples from 20 full-scale WWTPs. Nitrotoga-like bacteria were detected by PCR in 11 samples and reached abundances detectable by FISH in seven sludges. They coexisted with Nitrospira in most of these WWTPs, but constituted the only detectable NOB in two systems. Quantitative FISH revealed that Nitrotoga accounted for nearly 2% of the total bacterial community in one of these plants, a number comparable to Nitrospira abundances in other WWTPs. Spatial statistics revealed that Nitrotoga coaggregated with ammonia-oxidizing bacteria, strongly supporting a functional role in nitrite oxidation. This activity was confirmed by FISH in combination with microradiography, which revealed nitrite-dependent autotrophic carbon fixation by Nitrotoga in situ. Correlation of the presence or absence with WWTP operational parameters indicated low temperatures as a main factor supporting high Nitrotoga abundances, although in incubation experiments these NOB remained active over an unexpected range of temperatures, and also at different ambient nitrite concentrations. In conclusion, this study demonstrates that Nitrotoga can be functionally important nitrite oxidizers in WWTPs and can even represent the only known NOB in engineered systems.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E . (2007). Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J 1: 256–264.
Alawi M, Off S, Kaya M, Spieck E . (2009). Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep 1: 184–190.
Alm EW, Oerther DB, Larsen N, Stahl DA, Raskin L . (1996). The oligonucleotide probe database. Appl Environ Microbiol 62: 3557–3559.
Amann RI, Ludwig W, Schleifer KH . (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169.
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ . (2005). At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71: 7724–7736.
Behrens S, Ruhland C, Inacio J, Huber H, Fonseca A, Spencer-Martins I et al. (2003). In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl Environ Microbiol 69: 1748–1758.
Blöthe M, Roden EE . (2009). Composition and activity of an autotrophic Fe(II)-oxidizing, nitrate-reducing enrichment culture. Appl Environ Microbiol 75: 6937–6940.
Brümmer IH, Felske A, Wagner-Dobler I . (2003). Diversity and seasonal variability of Betaproteobacteria in biofilms of polluted rivers: analysis by temperature gradient gel electrophoresis and cloning. Appl Environ Microbiol 69: 4463–4473.
Camargo JA, Alonso Á . (2006). Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: a global assessment. Environ Int 32: 831–849.
Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H et al. (2009). Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J 3: 1093–1104.
Conley DJ, Paerl HW, Howarth RW, Boesch DF, Seitzinger SP, Havens KE et al. (2009). ECOLOGY: Controlling Eutrophication: nitrogen and phosphorus. Science 323: 1014–1015.
Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M . (1999). The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22: 434–444.
Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M . (2001a). In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 67: 5273–5284.
Daims H, Purkhold U, Bjerrum L, Arnold E, Wilderer PA, Wagner M . (2001b). Nitrification in sequencing biofilm batch reactors: lessons from molecular approaches. Water Sci Technol 43: 9–18.
Daims H, Stoecker K, Wagner M . (2005). Fluorescence in situ hybridization for the detection of prokaryotes. In: Osborn AM, Smith CJ (eds) Molecular Microbial Ecology. Taylor & Francis: New York, NY, USA, pp 213–239.
Daims H, Lücker S, Wagner M . (2006). daime, A novel image analysis program for microbial ecology and biofilm research. Environ Microbiol 8: 200–213.
Daims H, Wagner M . (2007). Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl Microbiol Biotechnol 75: 237–248.
Diaz RJ, Rosenberg R . (2008). Spreading dead zones and consequences for marine ecosystems. Science 321: 926–929.
Flynn TM, Sanford RA, Ryu H, Bethke CM, Levine AD, Ashbolt NJ et al. (2013). Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol 13: 146.
Foesel BU, Gieseke A, Schwermer C, Stief P, Koch L, Cytryn E et al. (2008). Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol Ecol 63: 192–204.
Garrity GM, Bell JA, Lilburn T . (2005). Family III. Gallionellaceae Henrici and Johnson 1935b. In: Brenner DY, Krieg NR, Staley JT (eds) Bergey's Manual of Systematic Bacteriology 2nd edn Springer: New York, NY, USA, pp 880–886.
Gujer W . (2010). Nitrification and me—a subjective review. Water Res 44: 1–19.
Henrici AT, Johnson DE . (1935). Studies of freshwater bacteria: II. Stalked bacteria, a new order of Schizomycetes. J Bacteriol 30: 61–93.
Henze M, Harremoes P, JlC Jansen, Arvin E . (1997) Wastwater Treatment—Biological and Chemical Processes 2 edn. Springer: Berlin, Germany.
Holm N . (2003). Process for the Discontinuous Purification and Installation for Carrying out this Process. European Patent Office, Germany, EP 0 834 474 B1.
Hoshino T, Yilmaz LS, Noguera DR, Daims H, Wagner M . (2008). Quantification of target molecules needed to detect microorganisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH. Appl Environ Microbiol 74: 5068–5077.
Ji Z, Chen Y . (2010). Using sludge fermentation liquid to improve wastewater short-cut nitrification-denitrification and denitrifying phosphorus removal via nitrite. Environ Sci Technol 44: 8957–8963.
Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Roser A, Koops HP et al. (1998). Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol 64: 3042–3051.
Kane MD, Poulsen LK, Stahl DA . (1993). Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol 59: 682–686.
Kong Y, Xia Y, Nielsen JL, Nielsen PH . (2007). Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiology 153: 4061–4073.
Kwon S, Kim TS, Yu GH, Jung JH, Park HD . (2010). Bacterial community composition and diversity of a full-scale integrated fixed-film activated sludge system as investigated by pyrosequencing. J Microbiol Biotechnol 20: 1717–1723.
Lebedeva EV, Alawi M, Maixner F, Jozsa PG, Daims H, Spieck E . (2008). Physiological and phylogenetic characterization of a novel lithoautotrophic nitrite-oxidizing bacterium, ‘Candidatus Nitrospira bockiana’. Int J Syst Evol Microbiol 58: 242–250.
Lee N, Nielsen PH, Andreasen KH, Juretschko S, Nielsen JL, Schleifer KH et al. (1999). Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol 65: 1289–1297.
Li D, Qi R, Yang M, Zhang Y, Yu T . (2011). Bacterial community characteristics under long-term antibiotic selection pressures. Water Res 45: 6063–6073.
Liu Z, Huang S, Sun G, Xu Z, Xu M . (2012). Phylogenetic diversity, composition and distribution of bacterioplankton community in the Dongjiang River, China. FEMS Microbiol Ecol 80: 30–44.
Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA 107: 13479–13484.
Ludwig W, Amann R, Martinez-Romero E, Schönhuber W, Bauer S, Neef A et al. (1998). rRNA based identification and detection systems for Rhizobia and other bacteria. Plant Soil 204: 1–19.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Maestre JP, Rovira R, Gamisans X, Kinney KA, Kirisits MJ, Lafuente J et al. (2009). Characterization of the bacterial community in a biotrickling filter treating high loads of H2S by molecular biology tools. Water Sci Technol 59: 1331–1337.
Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M et al. (2006). Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8: 1487–1495.
Maixner F, Wagner M, Lücker S, Pelletier E, Schmitz-Esser S, Hace K et al. (2008). Environmental genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium ‘Candidatus Nitrospira defluvii’. Environ Microbiol 10: 3043–3056.
Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC . (2011). Drivers of bacterial β-diversity depend on spatial scale. Proc Natl Acad Sci 108: 7850–7854.
Na H, Kim OS, Yoon SH, Kim Y, Chun J . (2011). Comparative approach to capture bacterial diversity of coastal waters. J Microbiol 49: 729–740.
Okabe S, Satoh H, Watanabe Y . (1999). In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol 65: 3182–3191.
Percent SF, Frischer ME, Vescio PA, Duffy EB, Milano V, McLellan M et al. (2008). Bacterial community structure of acid-impacted lakes: What controls diversity? Appl Environ Microbiol 74: 1856–1868.
Prosser JI . (1989). Autotrophic nitrification in bacteria. Adv Microb Physiol 30: 125–181.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596.
Ronquist F, Huelsenbeck JP . (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Sattin SR, Cleveland CC, Hood E, Reed SC, King AJ, Schmidt SK et al. (2009). Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition. J Microbiol 47: 673–681.
Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M et al. (2000). Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23: 93–106.
Schott J, Griffin BM, Schink B . (2010). Anaerobic phototrophic nitrite oxidation by Thiocapsa sp. strain KS1 and Rhodopseudomonas sp. strain LQ17. Microbiology 156: 2428–2437.
Schramm A, de Beer D, Wagner M, Amann R . (1998). Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol 64: 3480–3485.
Schramm A, de Beer D, van den Heuvel JC, Ottengraf S, Amann R . (1999). Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol 65: 3690–3696.
Schramm A, Fuchs BM, Nielsen JL, Tonolla M, Stahl DA . (2002). Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ Microbiol 4: 713–720.
Schwarz JI, Eckert W, Conrad R . (2007). Community structure of Archaea and Bacteria in a profundal lake sediment Lake Kinneret (Israel). Syst Appl Microbiol 30: 239–254.
Skerman VBD, McGowan V, Sneath PHA . (1980). Approved lists of bacterial names. Int J Syst Bacteriol 30: 225–420.
Sorokin DY, Vejmelkova D, Luecker S, Streshinskaya GM, Rijpstra I, Sinninghe Damsté J et al. (2014). Nitrolancea hollandica gen. nov., sp. nov., a chemolithoautotrophic nitrite-oxidizing bacterium from a bioreactor belonging to the phylum Chloroflexi. Int J Syst Evol Microbiol 64: 1859–1865.
Stein LY, Arp DJ . (1998). Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl Environ Microbiol 64: 4098–4102.
Tamminen M, Karkman A, Corander J, Paulin L, Virta M . (2011). Differences in bacterial community composition in Baltic Sea sediment in response to fish farming. Aquaculture 313: 15–23.
Wagner M, Rath G, Koops H-P, Flood J, Amann R . (1996). In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol 34: 237–244.
Wagner M, Loy A, Nogueira R, Purkhold U, Lee N, Daims H . (2002). Microbial community composition and function in wastewater treatment plants. Antonie Leeuwenhoek 81: 665–680.
Weiss J, Rentz J, Plaia T, Neubauer S, Merrill-Floyd M, Lilburn T et al. (2007). Characterization of neutrophilic Fe(II)-oxidizing bacteria isolated from the rhizosphere of wetland plants and description of Ferritrophicum radicicola gen. nov. sp. nov., and Sideroxydans paludicola sp. nov. Geomicrobiol J 24: 559–570.
White CP, Debry RW, Lytle DA . (2012). Microbial survey of a full-scale, biologically active filter for treatment of drinking water. Appl Environ Microbiol 78: 6390–6394.
Acknowledgements
We thank Anneliese Müller and Christian Baranyi for excellent technical assistance. Niels Holm, Dieter Schreff, Uwe Temper and the technical staff at the analyzed WWTPs are acknowledged for providing the activated sludge samples and for helpful discussions. This work was supported by the Austrian Science Fund (FWF) Grant P24101-B22, the Vienna Science and Technology Fund (WWTF) Grant LS09-040 and the German Research Foundation (DFG) Grant SP 667/3-1.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Data deposition: 16S rRNA gene sequence data have been submitted to the GenBank database under accession numbers KM017111–KM017129.
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Lücker, S., Schwarz, J., Gruber-Dorninger, C. et al. Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants. ISME J 9, 708–720 (2015). https://doi.org/10.1038/ismej.2014.158
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.158
This article is cited by
-
Diversity and distribution of iron-oxidising bacteria belonging to Gallionellaceae in different sites of a hydroelectric power plant
Brazilian Journal of Microbiology (2024)
-
The structure of microbial communities of activated sludge of large-scale wastewater treatment plants in the city of Moscow
Scientific Reports (2022)
-
Insights into the Effect of Sludge Retention Times on System Performance, Microbial Structure and Quorum Sensing in an Activated Sludge Bioreactor
Water, Air, & Soil Pollution (2022)
-
Relevance of Candidatus Nitrotoga for nitrite oxidation in technical nitrogen removal systems
Applied Microbiology and Biotechnology (2021)
-
Thickness determines microbial community structure and function in nitrifying biofilms via deterministic assembly
Scientific Reports (2019)