Abstract
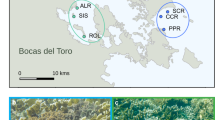
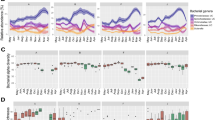
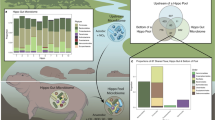
Diverse microbial consortia profoundly influence animal biology, necessitating an understanding of microbiome variation in studies of animal adaptation. Yet, little is known about such variability among fish, in spite of their importance in aquatic ecosystems. The Trinidadian guppy, Poecilia reticulata, is an intriguing candidate to test microbiome-related hypotheses on the drivers and consequences of animal adaptation, given the recent parallel origins of a similar ecotype across streams. To assess the relationships between the microbiome and host adaptation, we used 16S rRNA amplicon sequencing to characterize gut bacteria of two guppy ecotypes with known divergence in diet, life history, physiology and morphology collected from low-predation (LP) and high-predation (HP) habitats in four Trinidadian streams. Guts were populated by several recurring, core bacteria that are related to other fish associates and rarely detected in the environment. Although gut communities of lab-reared guppies differed from those in the wild, microbiome divergence between ecotypes from the same stream was evident under identical rearing conditions, suggesting host genetic divergence can affect associations with gut bacteria. In the field, gut communities varied over time, across streams and between ecotypes in a stream-specific manner. This latter finding, along with PICRUSt predictions of metagenome function, argues against strong parallelism of the gut microbiome in association with LP ecotype evolution. Thus, bacteria cannot be invoked in facilitating the heightened reliance of LP guppies on lower-quality diets. We argue that the macroevolutionary microbiome convergence seen across animals with similar diets may be a signature of secondary microbial shifts arising some time after host-driven adaptation.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR et al. (2011). Enterotypes of the human gut microbiome. Nature 473: 174–180.
Bassar RD, Marshall MC, López-Sepulcre A, Zandonà E, Auer SK, Travis J et al. (2010). Local adaptation in Trinidadian guppies alters ecosystem processes. Proc Natl Acad Sci USA 107: 3616–3621.
Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J et al. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 107: 18933–18938.
Bolnick DI, Snowberg LK, Gregory Caporaso J, Lauber C, Knight R, Stutz WE . (2014a). Major histocompatibility complex class IIb polymorphism influences gut microbiota composition and diversity. Mol Ecol 23: 4831–4845.
Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B et al. (2014b). Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun 5: 4500.
Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW . (2012). Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods 9: 425–426.
Cahill MM . (1990). Bacterial flora of fishes: a review. Microb Ecol 19: 21–41.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Chu C-C, Spencer JL, Curzi MJ, Zavala JA, Seufferheld MJ . (2013). Gut bacteria facilitate adaptation to crop rotation in the western corn rootworm. Proc Natl Acad Sci USA 110: 11917–11922.
Clemente Jose C, Ursell Luke K, Parfrey Laura W, Knight R . (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148: 1258–1270.
Clements KD, Angert ER, Montgomery WL, Choat JH . (2014). Intestinal microbiota in fishes: what’s known and what’s not. Mol Ecol 23: 1891–1898.
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 72: 5069–5072.
Douglas AE . (2009). The microbial dimension in insect nutritional ecology. Funct Ecol 23: 38–47.
El-Sabaawi RW, Zandonà E, Kohler TJ, Marshall MC, Moslemi JM, Travis J et al. (2012). Widespread intraspecific organismal stoichiometry among populations of the Trinidadian guppy. Funct Ecol 26: 666–676.
Endler JA . (1995). Multiple-trait coevolution and environmental gradients in guppies. Trends Ecol Evol 10: 22–29.
Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG et al. (2013). Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4: 1111–1119.
Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL et al. (2013). The long-term stability of the human gut microbiota. Science 341: 1237439.
Feldhaar H, Straka J, Krischke M, Berthold K, Stoll S, Mueller M et al. (2007). Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol 5: 48.
Fitzpatrick SW, Torres-Dowdall J, Reznick DN, Cameron KG, Funk WC . (2014). Parallelism isn’t perfect: could disease and flooding drive a life-history anomaly in Trinidadian guppies? AmNat 183: 290–300.
Flajnik MF, Du Pasquier L . (2004). Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol 25: 640–644.
Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG et al. (2013). Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci 110: E3730–E3738.
Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, Wilson ID et al. (2010). Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One 5: e8584.
Ghalambor CK, Reznick DN, Walker JA . (2004). Constraints on adaptive evolution: the functional trade-off between reproduction and fast-start swimming performance in the Trinidadian guppy (Poecilia reticulata). Am Nat 164: 38–50.
Grether GF, Millie DF, Bryant MJ, Reznick DN, Mayea W . (2001). Rain forest canopy cover, resource availability, and life history evolution in guppies. Ecology 82: 1546–1559.
Handelsman CA, Broder ED, Dalton CM, Ruell EW, Myrick CA, Reznick DN et al. (2013). Predator-induced phenotypic plasticity in metabolism and rate of growth: rapid adaptation to a novel environment. Integr Comp Biol 53: 975–988.
Herrel A, Huyghe K, Vanhooydonck B, Backeljau T, Breugelmans K, Grbac I et al. (2008). Rapid large-scale evolutionary divergence in morphology and performance associated with exploitation of a different dietary resource. Proc Natl Acad Sci USA 105: 4792–4795.
Hildebrand F, Nguyen TLA, Brinkman B, Yunta R, Cauwe B, Vandenabeele P et al. (2013). Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol 14: R4.
Hooper LV, Littman DR, Macpherson AJ . (2012). Interactions between the microbiota and the immune system. Science 336: 1268–1273.
Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T . (2007). Obligate symbiont involved in pest status of host insect. Proc R Soc B Biol Sci 274: 1979–1984.
Huse SM, Ye Y, Zhou Y, Fodor AA . (2012). A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7: e34242.
Jones RT, Sanchez LG, Fierer N . (2013). A cross-taxon analysis of insect-associated bacterial diversity. PLoS One 8: e61218.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M . (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114.
Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK et al. (2013). Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci USA 110: 17059–17064.
Kinnison M, Hendry A . (2001). The pace of modern life II: from rates of contemporary microevolution to pattern and process. In: Hendry AP, Kinnison MT, (eds) Microevolution Rate, Pattern, Process. Springer: The Netherlands, pp 145–164.
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotech 31: 814–821.
Lee YK, Mazmanian SK . (2010). Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330: 1768–1773.
Legendre P, Anderson MJ . (1999). Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol Monographs 69: 1–24.
Ley RE, Turnbaugh PJ, Klein S, Gordon JI . (2006). Human gut microbes associated with obesity. Nature 444: 1022–1023.
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS et al. (2008). Evolution of mammals and their gut microbes. Science 320: 1647–1651.
Linnenbrink M, Wang J, Hardouin EA, Künzel S, Metzler D, Baines JF . (2013). The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol 22: 1904–1916.
Loudon AH, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie V et al. (2014). Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J 8: 830–840.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Magurran AE . (2005) Evolutionary Ecology: The Trinidadian Guppy. Oxford University Press: New York.
McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA 110: 3229–3236.
Mountfort DO, Campbell J, Clements KD . (2002). Hindgut fermentation in three species of marine herbivorous fish. Appl Environ Microbiol 68: 1374–1380.
Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L et al. (2011). Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974.
Navarrete P, Magne F, Araneda C, Fuentes P, Barros L, Opazo R et al. (2012). PCR-TTGE analysis of 16S rRNA from Rainbow Trout (Oncorhynchus mykiss) gut microbiota reveals host-specific communities of active bacteria. PLoS One 7: e31335.
Nayak SK . (2010). Role of gastrointestinal microbiota in fish. Aquac Res 41: 1553–1573.
Nelson JS . (2006) Fishes of the World 4th edn. John Wiley & Sons, Inc.: Hoboken, New Jersey.
Nyholm SV, McFall-Ngai M . (2004). The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Micro 2: 632–642.
Ochman H, Worobey M, Kuo C-H, Ndjango J-BN, Peeters M, Hahn BH et al. (2010). Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8: e1000546.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2013). vegan: Community Ecology Package. R package version 2.0-8. Available at: http://CRAN.R-project.org/package=vegan.
R Development Core Team . (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing V: Austria, Available at: http://www.R-project.org/.
Rajilić-Stojanović M, Heilig HGHJ, Tims S, Zoetendal EG, de Vos WM . (2013). Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol 15: 1146–1159.
Rawls JF, Mahowald MA, Ley RE, Gordon JI . (2006). Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127: 423–433.
Reznick DN . (1983). The structure of guppy life histories: the tradeoff between growth and reproduction. Ecology 64: 862–873.
Reznick DN, Bryga HA . (1996). Life-history evolution in guppies (Poecilia reticulata: Poeciliidae). 5. Genetic basis of parallelism in life histories. Am Nat 147: 339–359.
Reznick DN, Rodd FH, Cardenas M . (1996). Life-history evolution in guppies (Poecilia reticulata: Poeciliidae). IV. Parallelism in life-history phenotypes. Am Nat 147: 319–338.
Reznick DN, Shaw FH, Rodd FH, Shaw RG . (1997). Evaluation of the rate of evolution in natural populations of guppies (Poecilia reticulata). Science 275: 1934–1937.
Reznick DN, Ricklefs RE . (2009). Darwin’s bridge between microevolution and macroevolution. Nature 457: 837–842.
Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341: 1241214.
Ringø E, Strøm E . (1994). Microflora of Arctic charr, Salvelinus alpinus (L.): gastrointestinal microflora of free-living fish and effect of diet and salinity on intestinal microflora. Aquac Res 25: 623–629.
Ringø E, Olsen RE . (1999). The effect of diet on aerobic bacterial flora associated with intestine of Arctic charr (Salvelinus alpinus L.). J Appl Microbiol 86: 22–28.
Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K et al. (2011). Evidence for a core gut microbiota in the zebrafish. ISME J 5: 1595–1608.
Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE . (2009). Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci USA 106: 21236–21241.
Sakata T, Sugita H, Mitsuoka T, Kakimoto D, Kadota H . (1981). Characteristics of obligate anaerobic bacteria in the intestines of freshwater fish. Bull Jap Soc Sci Fish 47: 421–427.
Sanders JG, Powell S, Kronauer DJC, Vasconcelos HL, Frederickson ME, Pierce NE . (2014). Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol Ecol 23: 1268–1283.
Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12: 277–288.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504.
Stamatakis A, Ludwig T, Meier H . (2005). RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21: 456–463.
Sugita H, Miyajima C, Deguchi Y . (1991). The vitamin B12-producing ability of the intestinal microflora of freshwater fish. Aquaculture 92: 267–276.
Sullam KE, Essinger SD, Lozupone CA, O’Connor MP, Rosen GL, Knight R et al. (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol 21: 3363–3378.
Sullam KE, Dalton CM, Russell JA, Kilham SS, El-Sabaawi R, German DP et al. (2014). Changes in digestive traits and body nutritional composition accommodate a trophic niche shift in Trinidadian guppies. Oecologia e-pub ahead of print 28 November 2014; doi:10.1007/s00442-014-3158-5.
Torres Dowdall J, Handelsman CA, Ruell EW, Auer SK, Reznick DN, Ghalambor CK . (2012). Fine-scale local adaptation in life histories along a continuous environmental gradient in Trinidadian guppies. Funct Ecol 26: 616–627.
Tremaroli V, Backhed F . (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489: 242–249.
Tsuchiya C, Sakata T, Sugita H . (2008). Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett Appl Microbiol 46: 43–48.
Van Soest PJ . (1994) Nutritional Ecology of the Ruminant 2nd edn. Cornell University Press: Ithaca, USA.
Willing E-M, Bentzen P, Van Oosterhout C, Hoffmann M, Cable J, Breden F et al. (2010). Genome-wide single nucleotide polymorphisms reveal population history and adaptive divergence in wild guppies. Mol Ecol 19: 968–984.
Wilson B, Danilowicz B, Meijer W . (2008). The diversity of bacterial communities associated with Atlantic Cod Gadus morhua. Microb Ecol 55: 425–434.
Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108.
Yang Z, Bielawski JP . (2000). Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15: 496–503.
Zandonà E . (2010) The trophic ecology of guppies (Poecilia reticulata) from the streams of Trinidad. Ph.D thesis, Drexel University: Philadelphia.
Zandonà E, Auer SK, Kilham SS, Howard JL, López-Sepulcre A, O’Connor MP et al. (2011). Diet quality and prey selectivity correlate with life histories and predation regime in Trinidadian guppies. Funct Ecol 25: 964–973.
Zilber-Rosenberg I, Rosenberg E . (2008). Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32: 723–735.
Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser JAGM, de Vos WM . (2001). The host genotype affects the bacterial community in the human gastronintestinal tract. Microbial Ecol Health Dis 13: 129–134.
Acknowledgements
We acknowledge funding from NSF-Frontiers in Integrative Biological Research grant (DEB-0623632EF) to ASF and an NSF-Doctoral Dissertation Improvement Grant (DEB- 1210695) to JAR, SSK and KES, a Sigma Xi Grant-in-aid to KES and the Betz Chair Endowment for funding this research. We thank Eugenia Zandonà for her invaluable assistance in the field, contribution of knowledge regarding field sites and helpful discussion. We also thank David Reznick and FIBR team for support, Keeley MacNeill for fieldwork assistance, Drew McQuade and Mauri Ren for laboratory assistance and Amy McCune for laboratory space at Cornell University. We thank two anonymous reviewers for their constructive comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Sullam, K., Rubin, B., Dalton, C. et al. Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of Trinidadian guppies. ISME J 9, 1508–1522 (2015). https://doi.org/10.1038/ismej.2014.231
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.231
This article is cited by
-
Geography and elevation as drivers of cloacal microbiome assemblages of a passerine bird distributed across Sulawesi, Indonesia
Animal Microbiome (2023)
-
Contrasting patterns of bacterial communities in the rearing water and gut of Penaeus vannamei in response to exogenous glucose addition
Marine Life Science & Technology (2022)
-
Flexibility and resilience of great tit (Parus major) gut microbiomes to changing diets
Animal Microbiome (2021)
-
Gut microbial communities associated with phenotypically divergent populations of the striped stem borer Chilo suppressalis (Walker, 1863)
Scientific Reports (2021)
-
European eel (Anguilla anguilla) GI tract conserves a unique metagenomics profile in the recirculation aquaculture system (RAS)
Aquaculture International (2021)