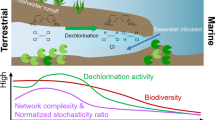
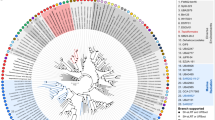
Abstract
The phylum Chloroflexi is one of the most frequently detected phyla in the subseafloor of the Pacific Ocean margins. Dehalogenating Chloroflexi (Dehalococcoidetes) was originally discovered as the key microorganisms mediating reductive dehalogenation via their key enzymes reductive dehalogenases (Rdh) as sole mode of energy conservation in terrestrial environments. The frequent detection of Dehalococcoidetes-related 16S rRNA and rdh genes in the marine subsurface implies a role for dissimilatory dehalorespiration in this environment; however, the two genes have never been linked to each other. To provide fundamental insights into the metabolism, genomic population structure and evolution of marine subsurface Dehalococcoidetes sp., we analyzed a non-contaminated deep-sea sediment core sample from the Peruvian Margin Ocean Drilling Program (ODP) site 1230, collected 7.3 m below the seafloor by a single cell genomic approach. We present for the first time single cell genomic data on three deep-sea Chloroflexi (Dsc) single cells from a marine subsurface environment. Two of the single cells were considered to be part of a local Dehalococcoidetes population and assembled together into a 1.38-Mb genome, which appears to be at least 85% complete. Despite a high degree of sequence-level similarity between the shared proteins in the Dsc and terrestrial Dehalococcoidetes, no evidence for catabolic reductive dehalogenation was found in Dsc. The genome content is however consistent with a strictly anaerobic organotrophic or lithotrophic lifestyle.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Adrian L, Reinhardt R, Kube M . (2009). Genomics as a tool to analyze bioremediation potentials and functional diversification in subsurface environments. Geochim Cosmochim Acta 73: A12.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Altschul SF, Koonin EV . (1998). Iterated profile searches with PSI-BLAST - a tool for discovery in protein databases. Trends Biochem Sci 23: 444–447.
Anderson I, Risso C, Holmes D, Lucas S, Copeland A, Lapidus A et al (2011). Complete genome sequence of Ferroglobus placidus AEDII12DO. Stand Genomic Sci 5: 50–60.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9: 75.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477.
Batzke A, Engelen B, Sass H, Cypionka H . (2007). Phylogenetic and physiological diversity of cultured deep-biosphere bacteria from equatorial Pacific Ocean and Peru Margin sediments. Geomicrobiol J 24: 261–273.
Berg IA . (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77: 1925–1936.
Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH . (2008). Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci USA 105: 10583–10588.
Biddle JF, Teske AP . (2008). A genetic view of diversity beneath the seafloor. Geochim Cosmochim Acta 72: A83.
Biddle JF, White JR, Teske AP, House CH . (2011). Metagenomics of the subsurface Brazos-Trinity Basin (IODP site 1320): comparison with other sediment and pyrosequenced metagenomes. ISME J 5: 1038–1047.
Blazejak A, Schippers A . (2010). High abundance of JS-1- and Chloroflexi-related Bacteria in deeply buried marine sediments revealed by quantitative, real-time PCR. FEMS Microbiol Ecol 72: 198–207.
Borowski WS, Paull CK, Ussler W . (1996). Marine pore-water sulfate profiles indicate in situ methane flux from underlying gas hydrate. Geology 24: 655–658.
Borowski WS, Hoehler TM, Alperin MJ, Rodriguez NM, Paull CK . (2000). Significance of anaerobic methane oxidation in methane-rich sediments overlying the Blake Ridge gas hydrates. In: Paull CK, Matsumoto R, Wallace PJ, Dillon WP (eds) Proceedings of the Ocean Drilling Program, Scientific Results vol. 164. Ocean Drilling Program: College Station, TX, pp 87–99.
Bryant DA, Liu ZF, Li T, Zhao FQ, Costas AMG, Klatt CG et al (2012). Comparative and functional genomics of anoxygenic green bacteria from the taxa Chlorobi, Chloroflexi, and Acidobacteria. Adv Photosynth Resp 33: 47–102.
Callaghan AV, Morris BEL, Pereira IAC, McInerney MJ, Austin RN, Groves JT et al (2012). The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation. Environ Microbiol 14: 101–113.
Cheng D, He J . (2009). Isolation and characterization of ‘Dehalococcoides’ sp. strain MB, which dechlorinates tetrachloroethene to trans-1,2-dichloroethene. Appl Environ Microbiol 75: 5910–5918.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145.
D’Hondt S, Jorgensen BB, Blake R, Dickens G, Hindrichs K, Holm N et al (2002a). Microbial activity in deeply buried marine sediments. Geochim Cosmochim Acta 66: A163.
D’Hondt S, Rutherford S, Spivack AJ . (2002b). Metabolic activity of subsurface life in deep-sea sediments. Science 295: 2067–2070.
D’Hondt S, Jørgensen BB, Miller DJ, The Leg 201 Scientific Party (2003a). ODP Leg 201 explores microbial life in deeply buried marine sediments off Peru. JOIDES J 29: 11–15.
D’Hondt S, Jorgensen BB, Miller DJ, Batzke A, Blake R, Cragg BA et al (2004). Distributions of microbial activities in deep subseafloor sediments. Science 306: 2216–2221.
D’Hondt SL, Jørgensen BB, Miller DJ . (2003b) Proceedings of the Ocean Drilling Program, Initial Reports, vol. 201. Ocean Drilling Program: College Station, TX, USA.
Durbin AM, Teske A . (2011). Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ Microbiol 13: 3219–3234.
Earl D, Bradnam K St, John J, Darling A, Lin DW, Fass J et al (2011). Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res 21: 2224–2241.
Fry JC, Parkes RJ, Cragg BA, Weightman AJ, Webster G . (2008). Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol Ecol 66: 181–196.
Futagami T, Morono Y, Terada T, Kaksonen AH, Inagaki F . (2009). Dehalogenation activities and distribution of reductive dehalogenase homologous genes in marine subsurface sediments. Appl Environ Microbiol 75: 6905–6909.
Futagami T, Morono Y, Terada T, Kaksonen AH, Inagaki F . (2013). Distribution of dehalogenation activity in subseafloor sediments of the Nankai Trough subduction zone. Philos Trans R Soc Lond B Biol Sci 368: 20120249.
He J, Sung Y, Krajmalnik-Brown R, Ritalahti KM, Löffler FE . (2005). Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ Microbiol 7: 1442–1450.
Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR et al (2013). Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1: 22.
Inagaki F, Suzuki M, Nealson KH, Horikoshi K, D’Hondt SL, Jorgensen BB et al (2003). Subseafloor microbial diversity in the Peru Margin (ODP Leg. 201). Geochim Cosmochim Acta 67: A171.
Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A et al (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments, on the Pacific Ocean Margin. Proc Natl Acad Sci USA 103: 2815–2820.
Iversen N, Jorgensen BB . (1985). Anaerobic methane oxidation rates at the sulfate methane transition in marine-sediments from Kattegat and Skagerrak (Denmark). Limnol Oceanogr 30: 944–955.
Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S . (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA 109: 16213–16216.
Kanehisa M, Goto S . (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30.
Kaster AK, Goenrich M, Seedorf H, Liesegang H, Wollherr A, Gottschalk G et al (2011a). More than 200 genes required for methane formation from H(2) and CO(2) and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus. Archaea 2011: 973848.
Kaster AK, Moll J, Parey K, Thauer RK . (2011b). Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci USA 108: 2981–2986.
Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA et al (1997). The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390: 364–370.
Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G et al (2012). Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 30: 693–700.
Krajmalnik-Brown R, Holscher T, Thomson IN, Saunders FM, Ritalahti KM, Loffler FE . (2004). Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol 70: 6347–6351.
Krzmarzick MJ, Crary BB, Harding JJ, Oyerinde OO, Leri AC, Myneni SCB et al (2012). Natural niche for organohalide-respiring Chloroflexi. Appl Environ Microbiol 78: 393–401.
Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, Adrian L . (2005). Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat Biotechnol 23: 1269–1273.
Kvenvolden KA, Frank TJ, Golan-Bac M . (1990). Hydrocarbon gases in tertiary and quaternary sediments offshore Peru—results and comparisons. In Suess E, von Huene R, et al. (eds) Proceedings of the Ocean Drilling Program, Scientific Results vol. 112. Ocean Drilling Program: College Station, TX, pp 505–515.
Kvenvolden KA, Kastner M . (1990). Gas hydrates of the Peruvian outer continental margin. In Suess E, von Huene R, et al. (eds) Proceedings of the Ocean Drilling Program, Scientific Results vol. 112. Ocean Drilling Program: College Station, TX, pp 517–526.
Lane D . (1991). 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic Acid Techniques in Bacterial Systematics. John Wiley: Chichester, UK.
Lasken RS . (2007). Single-cell genomic sequencing using Multiple Displacement Amplification. Curr Opin Microbiol 10: 510–516.
Lasken RS, Stockwell TB . (2007). Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol 7: 19.
Lasken RS . (2012). Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol 10: 631–640.
Lever MA, Rouxel O, Alt JC, Shimizu N, Ono SH, Coggon RM et al (2013). Evidence for microbial carbon and sulfur cycling in deeply buried ridge flank basalt. Science 339: 1305–1308.
Loeffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT et al (2013). Dehalococcoides mccartyi gen. nov., sp nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63: 625–635.
Marcy Y, Ishoey T, Lasken RS, Stockwell TB, Walenz BP, Halpern AL et al (2007). Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet 3: 1702–1708.
Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al (2010). The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res 38: D382–D390.
Mayer-Blackwell K, Spormann AM . (2014). Quantitative and massively parallel assessment of reductive dehalogenase genes. (submitted).
Maymo-Gatell X, Chien Y, Gossett JM, Zinder SH . (1997). Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276: 1568–1571.
McMurdie PJ, Behrens SF, Holmes S, Spormann AM . (2007). Unusual codon bias in vinyl chloride reductase genes of Dehalococcoides species. Appl Environ Microbiol 73: 2744–2747.
McMurdie PJ, Behrens SF, Müller JA, Göke J, Ritalahti KM, Wagner R et al (2009). Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet 5: e1000714.
McMurdie PJ, Hug LA, Edwards EA, Holmes S, Spormann AM . (2011). Site-specific mobilization of vinyl chloride respiration islands by a mechanism common in Dehalococcoides. BMC Genomics 12: 287.
Miller GS, Milliken CE, Sowers KR, May HD . (2005). Reductive dechlorination of tetrachloroethene to trans-dichloroethene and cis-dichloroethene by PCB-dechlorinating bacterium DF-1. Environ Sci Technol 39: 2631–2635.
Moe WM, Yan J, Nobre MF, da Costa MS, Rainey FA . (2009). Dehalogenimonas lykanthroporepellens gen. nov., sp. nov., a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater. Int J Syst Evol Micr 59: 2692–2697.
Muller JA, Rosner BM, von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM . (2004). Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp strain VS and its environmental distribution. Appl Environ Microbiol 70: 4880–4888.
Niewohner C, Hensen C, Kasten S, Zabel M, Schulz HD . (1998). Deep sulfate reduction completely mediated by anaerobic methane oxidation in sediments of the upwelling area off Namibia. Geochim Cosmochim Acta 62: 455–464.
Orcutt BN, Bach W, Becker K, Fisher AT, Hentscher M, Toner BM et al (2011). Colonization of subsurface microbial observatories deployed in young ocean crust. ISME J 5: 692–703.
Parkes RJ, Cragg BA, Wellsbury P . (2002). Recent studies on bacterial populations and processes in subseafloor sediments: a review (vol 8, pg 11, 2000). Hydrogeol J 10: 346–346.
Parkes RJ, Webster G, Cragg BA, Weightman AJ, Newberry CJ, Ferdelman TG et al (2005). Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 436: 390–394.
Poritz M, Goris T, Wubet T, Tarkka MT, Buscot F, Nijenhuis I et al (2013). Genome sequences of two dehalogenation specialists - Dehalococcoides mccartyi strains BTF08 and DCMB5 enriched from the highly polluted Bitterfeld region. FEMS Microbiol Lett 343: 101–104.
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C et al (2012). The Pfam protein families database. Nucleic Acids Res 40: D290–D301.
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF et al (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437.
Rozen S, Skaletski H . (2000). Primer3 on the WWW for general users and for biologist programmers. In Krawetz S, Misener S (eds) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press: Totowa, NJ, pp 365–386.
Shapiro HM . (2000). Microbial analysis at the single-cell level: tasks and techniques. J Microbiol Methods 42: 3–16.
Siddaramappa S, Challacombe JF, Delano SF, Green LD, Daligault H, Bruce D et al (2012). Complete genome sequence of Dehalogenimonas lykanthroporepellens type strain (BL-DC-9(T)) and comparison to ‘Dehalococcoides’ strains. Stand Genomic Sci 6: 251–264.
Sorokin DY, Lucker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, Rijpstra WIC et al (2012). Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J 6: 2245–2256.
Stojanowic A, Mander GJ, Duin EC, Hedderich R . (2003). Physiological role of the F-420-non-reducing hydrogenase (Mvh) from Methanothermobacter marburgensis. Arch Microbiol 180: 194–203.
Strittmatter AW, Liesegang H, Rabus R, Decker I, Amann J, Andres S et al (2009). Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ Microbiol 11: 1038–1055.
Suess E, von Huene R . (1988) Proceedings ofthe Ocean Drilling Program, Initial Reports, vol. 112. Ocean Drilling Program: College Station, TX, USA.
Suess E, von Huene R, Emeis KC, Burgois J, del G Cruzado J, De Wever P et al (1990) Proceedings of the Ocean Drilling Program, Scientific Results vol. 112. Ocean Drilling Program: College Station, TX, USA.
Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300.
Tas N, van Eekert MH, de Vos WM, Smidt H . (2010). The little bacteria that can - diversity, genomics and ecophysiology of ‘Dehalococcoides’ spp. in contaminated environments. Microb Biotechnol 3: 389–402.
Toffin L, Webster G, Weightman AJ, Fry JC, Prieur D . (2004). Molecular monitoring of culturable bacteria from deep-sea sediment of the Nankai Trough, Leg 190 Ocean Drilling Program. FEMS Microbiol Ecol 48: 357–367.
Valentine D . (2010). Emerging topics in marine methane biogeochemistry. Annu Rev Mar Sci 3: 147–171.
Vermeij P, van der Steen RJ, Keltjens JT, Vogels GD, Leisinger T . (1996). Coenzyme F390 synthetase from Methanobacterium thermoautotrophicum Marburg belongs to the superfamily of adenylate-forming enzymes. J Bacteriol 178: 505–510.
Vermeij P, Pennings JL, Maassen SM, Keltjens JT, Vogels GD . (1997). Cellular levels of factor 390 and methanogenic enzymes during growth of Methanobacterium thermoautotrophicum deltaH. J Bacteriol 179: 6640–6648.
Wagner A, Cooper M, Ferdi S, Seifert J, Adrian L . (2012). Growth of Dehalococcoides mccartyi strain CBDB1 by reductive dehalogenation of brominated benzenes to benzene. Environ Sci Technol 46: 8960–8968.
Wasmund K, Schreiber L, Lloyd KG, Petersen DG, Schramm A, Stepanauskas R et al (2013). Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi. ISME J 8: 383–397.
Webster G, Parkes RJ, Fry JC, Weightman AJ . (2004). Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl Environ Microbiol 70: 5708–5713.
Webster G, Parkes RJ, Cragg BA, Newberry CJ, Weightman AJ, Fry JC . (2006). Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol Ecol 58: 65–85.
Webster G, Sass H, Cragg BA, Gorra R, Knab NJ, Green CJ et al (2011). Enrichment and cultivation of prokaryotes associated with the sulphate-methane transition zone of diffusion-controlled sediments of Aarhus Bay, Denmark, under heterotrophic conditions. FEMS Microbiol Ecol 77: 248–263.
Woyke T, Sczyrba A, Lee J, Rinke C, Tighe D, Clingenpeel S et al (2011). Decontamination of MDA reagents for single cell whole genome amplification. PLoS ONE 6: e26161.
Yamada T, Sekiguchi Y . (2009). Cultivation of uncultured chloroflexi subphyla: significance and ecophysiology of formerly uncultured Chloroflexi ‘Subphylum I’ with natural and biotechnological relevance. Microbes Environ 24: 205–216.
Zhu F, Massana R, Not F, Marie D, Vaulot D . (2005). Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52: 79–92.
Acknowledgements
This project was funded by the NSF Center for Dark Energy Biosphere Investigations (C-DEBI) and a fellowship to AK by C-DEBI and the Deutsche Forschungsgemeinschaft. We thank Jennifer Biddle (UC Delaware), Taiki Futagami and Fumio Inagaki (Kochi Institute for Core sample Research, Japan) for deep-sea sediment samples. We thank Ramunas Stepanauskas from the Bigelow Laboratory Single Cell Genomics Center for single cell sorting. We also thank Sergey Koren from the National Biodefense Analysis and Countermeasures Center for bioinformatics support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Kaster, AK., Mayer-Blackwell, K., Pasarelli, B. et al. Single cell genomic study of Dehalococcoidetes species from deep-sea sediments of the Peruvian Margin. ISME J 8, 1831–1842 (2014). https://doi.org/10.1038/ismej.2014.24
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.24
Keywords
This article is cited by
-
Microbial debromination of hexabromocyclododecanes
Applied Microbiology and Biotechnology (2021)
-
Microbial community and geochemical analyses of trans-trench sediments for understanding the roles of hadal environments
The ISME Journal (2020)
-
Microbial single-cell omics: the crux of the matter
Applied Microbiology and Biotechnology (2020)
-
Methane-fuelled biofilms predominantly composed of methanotrophic ANME-1 in Arctic gas hydrate-related sediments
Scientific Reports (2019)
-
Ecology and evolution of seafloor and subseafloor microbial communities
Nature Reviews Microbiology (2018)