Abstract
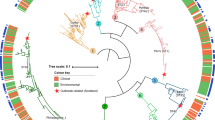
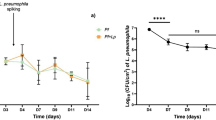
It is generally accepted that selection for resistance to grazing by protists has contributed to the evolution of Legionella pneumophila as a pathogen. Grazing resistance is becoming more generally recognized as having an important role in the ecology and evolution of bacterial pathogenesis. However, selection for grazing resistance presupposes the existence of protist grazers that provide the selective pressure. To determine whether there are protists that graze on pathogenic Legionella species, we investigated the existence of such organisms in a variety of environmental samples. We isolated and characterized diverse protists that graze on L. pneumophila and determined the effects of adding L. pneumophila on the protist community structures in microcosms made from these environmental samples. Several unrelated organisms were able to graze efficiently on L. pneumophila. The community structures of all samples were markedly altered by the addition of L. pneumophila. Surprisingly, some of the Legionella grazers were closely related to species that are known hosts for L. pneumophila, indicating the presence of unknown specificity determinants for this interaction. These results provide the first direct support for the hypothesis that protist grazers exert selective pressure on Legionella to acquire and retain adaptations that contribute to survival, and that these properties are relevant to the ability of the bacteria to cause disease in people. We also report a novel mechanism of killing of amoebae by one Legionella species that requires an intact Type IV secretion system but does not involve intracellular replication. We refer to this phenomenon as ‘food poisoning’.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR et al. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol 52: 399–451.
Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS et al. (2012). The revised classification of eukaryotes. J Eukaryot Microbiol 59: 429–493.
Al-Quadan T, Price CT, Abu Kwaik Y . (2012). Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol 20: 299–306.
Anderson OR, Wang W, Faucher SP, Bi K, Shuman HA . (2011). A new Heterolobosean amoeba Solumitrus palustris n. g., n. sp. isolated from freshwater marsh soil. J Eukaryot Microbiol 58: 60–67.
Bell T, Bonsall MB, Buckling A, Whiteley AS, Goodall T, Griffiths RI . (2010). Protists have divergent effects on bacterial diversity along a productivity gradient. Biol Lett 6: 639–642.
Berger KH, Isberg RR . (1993). Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7: 7–19.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108 (Suppl 1): 4516–4522.
Cavalier-Smith T . (2002). The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52: 297–354.
Cavalier-Smith T, Oates B . (2012). Ultrastructure of Allapsa vibrans and the body plan of Glissomonadida (Cercozoa). Protist 163: 165–187.
Davies B, Chattings LS, Edwards SW . (1991). Superoxide generation during phagocytosis by Acanthamoeba castellanii: similarities to the respiratory burst of immune phagocytes. Microbiology 137: 705–710.
Declerck P, Behets J, Delaedt Y, Margineanu A, Lammertyn E, Ollevier F . (2005). Impact of non-Legionella bacteria on the uptake and intracellular replication of Legionella pneumophila in Acanthamoeba castellanii and Naegleria lovaniensis. Microb Ecol 50: 536–549.
Dey R, Bodennec J, Mameri MO, Pernin P . (2009). Free-living freshwater amoebae differ in their susceptibility to the pathogenic bacterium Legionella pneumophila. FEMS Microbiol Lett 290: 10–17.
Edelstein PH, Edelstein MA, Shephard LJ, Ward KW, Ratcliff RM . (2012). Legionella steelei sp. nov., isolated from human respiratory specimens in California, USA, and South Australia. Int J Syst Evol Microbiol 62: 1766–1771.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R . (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200.
Ensminger AW, Isberg RR . (2009). Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr Opin Microbiol 12: 67–73.
Fields BS . (1996). The molecular ecology of Legionellae. Trends Microbiol 4: 286–290.
Fouque E, Trouilhe MC, Thomas V, Hartemann P, Rodier MH, Hechard Y . (2012). Cellular, biochemical, and molecular changes during encystment of free-living amoebae. Eukaryotic Cell 11: 382–387.
Franco IS, Shuman HA, Charpentier X . (2009). The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol 11: 1435–1443.
Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG et al. (1977). Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med 297: 1189–1197.
Gao LY, Harb OS, Abu Kwaik Y . (1997). Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect Immun 65: 4738–4746.
Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C . (2011). Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front Microbiol 2: 208.
Hahn MW, Hofle MG . (2001). Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol Eco 35: 113–121.
Jousset A . (2012). Ecological and evolutive implications of bacterial defences against predators. Environ Microbiol 14: 1830–1843.
Kilvington S, Price J . (1990). Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J Appl Bacteriol 68: 519–525.
Knodler LA, Celli J . (2011). Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell Microbiol 13: 1319–1327.
Lamoth F, Greub G . (2010). Amoebal pathogens as emerging causal agents of pneumonia. FEMS Microbiol Rev 34: 260–280.
Luo ZQ . (2012). Legionella secreted effectors and innate immune responses. Cell Microbiol 14: 19–27.
Levine B, Mizushima N, Virgin HW . (2011). Autophagy in immunity and inflammation. Nature 469: 323–335.
Matz C, Kjelleberg S . (2005). Off the hook—how bacteria survive protozoan grazing. Trends Microbiol 13: 302–307.
McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR . (1977). Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med 297: 1197–1203.
Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y . (2005). Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol 71: 20–28.
Nash TW, Libby DM, Horwitz MA . (1984). Interaction between the legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J Clin Invest 74: 771–782.
Nash TW, Libby DM, Horwitz MA . (1988). IFN-gamma-activated human alveolar macrophages inhibit the intracellular multiplication of Legionella pneumophila. J Immunol 140: 3978–3981.
Newsome AL, Baker RL, Miller RD, Arnold RR . (1985). Interactions between Naegleria fowleri and Legionella pneumophila. Infect Immun 50: 449–452.
Ninio S, Roy CR . (2007). Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol 15: 372–380.
O'Connor TJ, Adepoju Y, Boyd D, Isberg RR . (2011). Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci USA 108: 14733–14740.
Page FC . (1988) A New Key to Freshwater and Soil Gymnamoebae With Instructions for Culture. Freshwater Biological Association: Cumbria.
Pernthaler J . (2005). Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3: 537–546.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596.
Rowbotham TJ . (1980). Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol 33: 1179–1183.
Segal G, Purcell M, Shuman HA . (1998). Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA 95: 1669–1674.
Segal G, Shuman HA . (1999). Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun 67: 2117–2124.
Segal G . (2013). Identification of Legionella effectors using bioinformatic approaches. Methods Mol Biol 954: 595–602.
Shevchuk O, Jager J, Steinert M . (2011). Virulence properties of the Legionella pneumophila cell envelope. Front Microbiol 2: 74.
Solomon JM, Rupper A, Cardelli JA, Isberg RR . (2000). Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect Immun 68: 2939–2947.
Stoeck T, Zuendorf A, Breiner HW, Behnke A . (2007). A molecular approach to identify active microbes in environmental eukaryote clone libraries. Microb Ecol 53: 328–339.
Valster RM, Wullings BA, van der Kooij D . (2010). Detection of protozoan hosts for Legionella pneumophila in engineered water systems by using a biofilm batch test. Appl Environ Microb 76: 7144–7153.
Vogel JP, Andrews HL, Wong SK, Isberg RR . (1998). Conjugative transfer by the virulence system of Legionella pneumophila. Science 279: 873–876.
Acknowledgements
This work was supported by an award from NIAID, 5RO1AI23549 (H.A.S.) and funds from the Department of Microbiology, University of Chicago. F Amaro was supported by a postdoctoral fellowship from the Fulbright Commission and Ministry of Education of Spain. We thank our colleagues for supplying samples of various Legionella species, including Barry Fields and Claressa Lucas at CDC and Paul Edelstein at the University of Pennsylvania. We would like to thank the following individuals for supplying environmental samples used in this work: Dr Juan Carlos Gutiérrez from the Department of Microbiology III, Complutense University of Madrid, Dr Maureen Coleman from the Department of Geophysics at the University of Chicago and Dr Geeta K Rijal from the Metropolitan Water Reclamation District of Greater Chicago. The authors would like to express their appreciation to the Integrated Ocean Drilling Program shipboard technical, operations and engineering personnel of Integrated Ocean Drilling Program Expedition 311 for collecting, archiving and providing the ocean subsurface sample used in this research. Funding for Integrated Ocean Drilling Program was provided by the following agencies at the time of this expedition: European Consortium for Ocean Research Drilling, Ministry of Education, Culture, Sports, Science and Technology of Japan. Ministry of Science and Technology, People’s Republic of China, National Science Foundation (United States). Any opinions, findings and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the participating agencies, Integrated Ocean Drilling Program Management International or the Integrated Ocean Drilling Program Implementing Organizations. This is Lamont–Doherty Contribution No. 7850.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Amaro, F., Wang, W., Gilbert, J. et al. Diverse protist grazers select for virulence-related traits in Legionella. ISME J 9, 1607–1618 (2015). https://doi.org/10.1038/ismej.2014.248
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.248
This article is cited by
-
Hyphosphere microbiome of arbuscular mycorrhizal fungi: a realm of unknowns
Biology and Fertility of Soils (2023)
-
Microbial warfare in the wild—the impact of protists on the evolution and virulence of bacterial pathogens
International Microbiology (2021)
-
Unravelling the importance of the eukaryotic and bacterial communities and their relationship with Legionella spp. ecology in cooling towers: a complex network
Microbiome (2020)
-
Combinatorial selection in amoebal hosts drives the evolution of the human pathogen Legionella pneumophila
Nature Microbiology (2020)
-
Burkholderia bacteria use chemotaxis to find social amoeba Dictyostelium discoideum hosts
The ISME Journal (2018)