Abstract
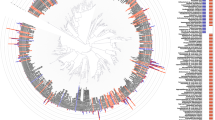
Determining the composition and function of subgingival dental plaque is crucial to understanding human periodontal health and disease, but it is challenging because of the complexity of the interactions between human microbiomes and human body. Here, we examined the phylogenetic and functional gene differences between periodontal and healthy individuals using MiSeq sequencing of 16S rRNA gene amplicons and a specific functional gene array (a combination of GeoChip 4.0 for biogeochemical processes and HuMiChip 1.0 for human microbiomes). Our analyses indicated that the phylogenetic and functional gene structure of the oral microbiomes were distinctly different between periodontal and healthy groups. Also, 16S rRNA gene sequencing analysis indicated that 39 genera were significantly different between healthy and periodontitis groups, and Fusobacterium, Porphyromonas, Treponema, Filifactor, Eubacterium, Tannerella, Hallella, Parvimonas, Peptostreptococcus and Catonella showed higher relative abundances in the periodontitis group. In addition, functional gene array data showed that a lower gene number but higher signal intensity of major genes existed in periodontitis, and a variety of genes involved in virulence factors, amino acid metabolism and glycosaminoglycan and pyrimidine degradation were enriched in periodontitis, suggesting their potential importance in periodontal pathogenesis. However, the genes involved in amino acid synthesis and pyrimidine synthesis exhibited a significantly lower relative abundance compared with healthy group. Overall, this study provides new insights into our understanding of phylogenetic and functional gene structure of subgingival microbial communities of periodontal patients and their importance in pathogenesis of periodontitis.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Abe K . (2012). Butyric acid induces apoptosis in both human monocytes and lymphocytes equivalently. J Oral Sci 54: 7–14.
Abusleme L, Dupuy A, Dutzan N, Silva N, Burleson J, Strausbaugh L et al (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J 7: 1016–1025.
Aimetti M, Cacciatore S, Graziano A, Tenori L . (2012). Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics 8: 465–474.
Alonso De La Peña V, Diz Dios P, Tojo Sierra R . (2007). Relationship between lactate dehydrogenase activity in saliva and oral health status. Arch Oral Biol 52: 911–915.
Amano A . (2010). Bacterial adhesins to host components in periodontitis. Periodontol 2000 52: 12–37.
Anderson MJ . (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
Armitage GC . (1999). Development of a classification system for periodontal diseases and conditions. Ann Periodontol 4: 1–6.
Armitage GC . (2004). Analysis of gingival crevice fluid and risk of progression of periodontitis. Periodontol 2000 34: 109–119.
Aruni AW, Roy F, Fletcher HM . (2011). Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun 79: 3872–3886.
Barnes VM, Teles R, Trivedi HM, Devizio W, Xu T, Mitchell MW et al (2009). Acceleration of purine degradation by periodontal diseases. J Dent Res 88: 851–855.
Barnes VM, Ciancio SG, Shibly O, Xu T, Devizio W, Trivedi HM et al (2011). Metabolomics reveals elevated macromolecular degradation in periodontal disease. J Dent Res 90: 1293–1297.
Braun V, Focareta T . (1991). Pore-forming bacterial protein hemolysins (cytolysins). Crit Rev Microbiol 18: 115–158.
Chang MC, Tsai YL, Chen YW, Chan CP, Huang CF, Lan WC et al (2013). Butyrate induces reactive oxygen species production and affects cell cycle progression in human gingival fibroblasts. J Periodontal Res 48: 66–73.
Clarke KR . (1993). Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18: 117–143.
Colombo APV, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD et al (2009). Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the Human Oral Microbe Identification Microarray. J Periodontol 80: 1421–1432.
Colombo APV, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD et al (2012). Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol 83: 1279–1287.
Cossart P, Jonquières R . (2000). Sortase, a universal target for therapeutic agents against Gram-positive bacteria? Proc Natl Acad Sci USA 97: 5013–5015.
Darveau RP . (2010). Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Micro 8: 481–490.
de Hoon MJL, Imoto S, Nolan J, Miyano S . (2004). Open source clustering software. Bioinformatics 20: 1453–1454.
Dietrich T, Jimenez M, Krall Kaye EA, Vokonas PS, Garcia RI . (2008). Age-dependent associations between chronic periodontitis/edentulism and risk of coronary heart disease. Circulation 117: 1668–1674.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Eley BM, Cox SW . (2003). Proteolytic and hydrolytic enzymes from putative periodontal pathogens: characterization, molecular genetics, effects on host defenses and tissues and detection in gingival crevice fluid. Periodontol 2000 31: 105–124.
Ertugrul AS, Arslan U, Dursun R, Hakki SS . (2013). Periodontopathogen profile of healthy and oral lichen planus patients with gingivitis or periodontitis. Int J Oral Sci 5: 92–97.
Fischer CL, Walters KS, Drake DR, Dawson DV, Blanchette DR, Brogden KA et al (2013). Oral mucosal lipids are antibacterial against Porphyromonas gingivalis, induce ultrastructural damage, and alter bacterial lipid and protein compositions. Int J Oral Sci 5: 130–140.
Giannobile WV, Al-Shammari KF, Sarment DP . (2003). Matrix molecules and growth factors as indicators of periodontal disease activity. Periodontol 2000 31: 125–134.
Graves DT, Jiang Y, Genco C . (2000). Periodontal disease: bacterial virulence factors, host response and impact on systemic health. Curr Opin Infect Dis 13: 227–232.
Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK et al (2012a). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6: 1176–1185.
Hajishengallis G, Lamont RJ . (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 27: 409–419.
He XS, Shi WY . (2009). Oral microbiology: past, present and future. Int J Oral Sci 1: 47–58.
He Z, Nostrand J, Deng Y, Zhou J . (2011). Development and applications of functional gene microarrays in the analysis of the functional diversity, composition, and structure of microbial communities. Front Environ Sci Engin China 5: 1–20.
He Z, Deng Y, Zhou J . (2012a). Development of functional gene microarrays for microbial community analysis. Curr Opin Biotechnol 23: 49–55.
He Z, Van Nostrand JD, Zhou J . (2012b). Applications of functional gene microarrays for profiling microbial communities. Curr Opin Biotechnol 23: 460–466.
Holt SC, Ebersole JL . (2005). Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000 38: 72–122.
Kuboniwa M, Hendrickson E, Xia Q, Wang T, Xie H, Hackett M et al (2009). Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol 9: 98.
Kumar PS, Griffen AL, Moeschberger ML, Leys EJ . (2005). Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol 43: 3944–3955.
Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL . (2006). Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol 44: 3665–3673.
Last KS, Stanbury JB, Embery G . (1985). Glycosaminoglycans in human gingival crevicular fluid as indicators of active periodontal disease. Arch Oral Biol 30: 275–281.
Letunic I, Bork P . (2011). Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478.
Liu B, Faller L, Klitgord N, Mazumdar V, Ghodsi M, Sommer D et al (2012). Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One 7: e37919.
Lu RF, Meng HX, Gao XJ, Feng L, Xu L . (2008). Analysis of short chain fatty acids in gingival crevicular fluid of patients with aggressive periodontitis. Chinese J Stomatol 43: 664–667.
Marx JJM . (2002). Iron and infection: competition between host and microbes for a precious element. Best Pract Res Cl Ha 15: 411–426.
Mazumdar V, Snitkin ES, Amar S, Segrè D . (2009). Metabolic network model of a human oral pathogen. J Bacteriol 191: 74–90.
Meng H . (2008) Periodontology. People’s Medical Publishing House: Peking.
Moffatt CE, Whitmore SE, Griffen AL, Leys EJ, Lamont RJ . (2011). Filifactor alocis interactions with gingival epithelial cells. Mol Oral Microbiol 26: 365–373.
Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA et al (2003). Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis Strain W83. J Bacteriol 185: 5591–5601.
Petersen PE, Bourgeois D, Ogawa H, Estupinan-Day S, Ndiaye C . (2005). The global burden of oral diseases and risks to oral health. Bull World Health Organ 83: 661–669.
Pihlstrom BL, Michalowicz BS, Johnson NW . (2005). Periodontal diseases. Lancet 366: 1809–1820.
Qiqiang L, Huanxin M, Xuejun G . (2012). Longitudinal study of volatile fatty acids in the gingival crevicular fluid of patients with periodontitis before and after nonsurgical therapy. J Periodontal Res 47: 740–749.
Rai B, Kharb S, Jain R, Anand SC . (2008). Biomarkers of periodontitis in oral fluids. J Oral Sci 50: 53–56.
Rakoto-Alson S, Tenenbaum H, Davideau J-L . (2009). Periodontal diseases, preterm births, and low birth weight: findings from a homogeneous cohort of women in Madagascar. J Periodontol 81: 205–213.
Sandholm L . (1986). Proteases and their inhibitors in chronic inflammatory periodontal disease. J Clin Periodontol 13: 19–26.
Sfyroeras GS, Roussas N, Saleptsis VG, Argyriou C, Giannoukas AD . (2012). Association between periodontal disease and stroke. J Vasc Surg 55: 1178–1184.
Smith AJ, Addy M, Embery G . (1995). Gingival crevicular fluid glycosaminoglycan levels in patients with chronic adult periodontitis. J Clin Periodontol 22: 355–361.
Socransky SS, Haffajee AD . (1992). The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol 63: 322–331.
Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL . (1998). Microbial complexes in subgingival plaque. J Clin Periodontol 25: 134–144.
Stehle HW, Leblebicioglu B, Walters JD . (2001). Short-chain carboxylic acids produced by gram-negative anaerobic bacteria can accelerate or delay Polymorphonuclear leukocyte apoptosis in vitro. J Periodontol 72: 1059–1063.
Suvan J, D'Aiuto F, Moles DR, Petrie A, Donos N . (2011). Association between overweight/obesity and periodontitis in adults. A systematic review. Obes Rev 12: 381–404.
Takahashi N, Sato T . (2002). Dipeptide utilization by the periodontal pathogens Porphyromonas gingivalis, Prevotella intermedia, Prevotella nigrescens and Fusobacterium nucleatum. Oral Microbiol Immunol 17: 50–54.
Tanner ACR, Kent R, Kanasi E, Lu SC, Paster BJ, Sonis ST et al (2007). Clinical characteristics and microbiota of progressing slight chronic periodontitis in adults. J Clin Periodontol 34: 917–930.
Tatakis DN, Kumar PS . (2005). Etiology and pathogenesis of periodontal diseases. Dent Clin North Am 49: 491–516.
Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G . (2006). Pili in Gram-positive pathogens. Nat Rev Micro 4: 509–519.
Tonetti MS . (2009). Periodontitis and risk for atherosclerosis: an update on intervention trials. J Clin Periodontol 36: 15–19.
Tsuda H, Ochiai K, Suzuki N, Otsuka K . (2010). Butyrate, a bacterial metabolite, induces apoptosis and autophagic cell death in gingival epithelial cells. J Periodontal Res 45: 626–634.
Tu Q, He Z, Li Y, Chen Y, Deng Y, Lin L et al (2014b). Development of HuMiChip for Functional Profiling of Human Microbiomes. PLoS One 9: e90546.
Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD et al (2014a). GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour e-pub ahead of print 12 February 2014 doi:10.1111/1755-0998.12239.
Waddington RJ, Embery G, Last KS . (1989). Glycosaminoglycans of human alveolar bone. Arch Oral Biol 34: 587–589.
Acknowledgements
This work was supported by the International Science and Technology Cooperation Program of China (Grant Number: 2011DFA30940), the National Basic Research Program of China (‘973 Pilot Research Program’, Grant Number: 2011CB512108), National Key Technologies R&D Program of the Twelve Five-Year Plan, the Ministry of Science and Technology of China (Grant 2012BAI07B03), the National Natural Science Foundation of China (81170959, 30901689 and 81172579), Sichuan Provincial Department of science and technology project (2013SZ0039) and by Oklahoma Applied Research Support (OARS), Oklahoma Center for the Advancement of Science and Technology (OCAST), the State of Oklahoma through the Project AR11-035. We are also grateful to Tong Yuan, Xiaofei Lu, Feifei Liu, Qichao Tu, Zhou Shi, Kai Xue, Lei Cheng (OU researchers) and Arthur Escalas (PhD student of Université Montpellier) for their great help on this project.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Li, Y., He, J., He, Z. et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J 8, 1879–1891 (2014). https://doi.org/10.1038/ismej.2014.28
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.28
Keywords
This article is cited by
-
Enhanced propagation of Granulicatella adiacens from human oral microbiota by hyaluronan
Scientific Reports (2022)
-
Anna Karenina and the subgingival microbiome associated with periodontitis
Microbiome (2021)
-
Beware of pharyngeal Fusobacterium nucleatum in COVID-19
BMC Microbiology (2021)
-
The role of the oral microbiota in chronic non-communicable disease and its relevance to the Indigenous health gap in Australia
BMC Oral Health (2020)
-
The subgingival microbiome associated with periodontitis in type 2 diabetes mellitus
The ISME Journal (2020)