Abstract
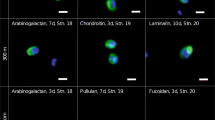
We compared the function and composition of free-living and particle-associated microbial communities at an inshore site in coastal North Carolina and across a depth profile on the Blake Ridge (offshore). Hydrolysis rates of six different polysaccharide substrates were compared for particle-associated (>3 μm) and free-living (<3 to 0.2 μm) microbial communities. The 16S rRNA- and rDNA-based clone libraries were produced from the same filters used to measure hydrolysis rates. Particle-associated and free-living communities resembled one another; they also showed similar enzymatic hydrolysis rates and substrate preferences. All six polysaccharides were hydrolyzed inshore. Offshore, only a subset was hydrolyzed in surface water and at depths of 146 and 505 m; just three polysaccharides were hydrolyzed at 505 m. The spectrum of bacterial taxa changed more subtly between inshore and offshore surface waters, but changed greatly with depth offshore. None of the OTUs occurred at all sites: 27 out of the 28 major OTUs defined in this study were found either exclusively in a surface or in a mid-depth/bottom water sample. This distinction was evident with both 16S rRNA and rDNA analyses. At the offshore site, despite the low community overlap, bacterial communities maintained a degree of functional redundancy on the whole bacterial community level with respect to hydrolysis of high-molecular-weight substrates.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Acinas S, Anton J, Rodriguez-Valera F . (1999). Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol 65: 514–522.
Agogué H, Lamy D, Neal PR, Sogin ML, Herndl GJ . (2011). Water mass–specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol Ecol 20: 258–274.
Alderkamp AC, Van Rijssel M, Bolhuis H . (2007). Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol Ecol 59: 108–117.
Alldredge AL, Cole JJ, Caron DA . (1986). Production of heterotrophic bacteria inhabiting macroscopic organic aggregates (marine snow) from surface waters. Limnol Oceanogr 31: 68–78.
Alldredge AL, Gotschalk CC . (1990). The relative contribution of marine snow of different origins to biological processes in coastal waters. Cont Shelf Res 10: 41–58.
Alldredge AL, Passow U, Logan BE . (1993). The abundance and significance of a class of large, transparent organic particles in the ocean. Deep Sea Res I 40: 1131–1140.
Alonso C, Warnecke F, Amann R, Pernthaler J . (2007). High local and global diversity of Flavobacteria in marine plankton. Environ Microbiol 9: 1253–1266.
Alonso-Sáez L, Arístegui J, Pinhassi J, Gómez-Consarnau L, González JM, Vaqué D et al. (2006). Bacterial assemblage structure and carbon metabolism along a productivity gradient in the NE Atlantic Ocean. Aquat Microb Ecol 46: 43–53.
Amon RMW, Fitznar HP, Benner R . (2001). Linkages among the bioreactivity, chemical composition, and diagenetic state of marine dissolved organic matter. Limnol Oceanogr 46: 287–297.
Arnosti C . (1995). Measurement of depth- and site-related differences in polysaccharide hydrolysis rates in marine sediments. Geochim Cosmochim Acta 59: 4247–4257.
Arnosti C . (2003). Fluorescent derivatization of polysaccharides and carbohydrate-containing biopolymers for measurement of enzyme activities in complex media. J Chromatogr B 793: 181–191.
Arnosti C . (2004). Speed bumps and barricades in the carbon cycle: substrate structural effects on carbon cycling. Mar Chem 92: 263–273.
Arnosti C . (2008). Functional differences between Arctic seawater and sedimentary microbial communities: contrasts in microbial hydrolysis of complex substrates. FEMS Microbiol Ecol 66: 343–351.
Arnosti C . (2011). Microbial extracellular enzymes and the marine carbon cycle. Ann Rev Mar Sci 3: 401–425.
Arnosti C, Durkin S, Jeffrey WH . (2005). Patterns of extracellular enzyme activities among pelagic marine microbial communities: implications for cycling of dissolved organic carbon. Aquat Microb Ecol 38: 135–145.
Arnosti C, Ziervogel K, Ocampo L, Ghobrial S . (2009). Enzyme activities in the water column and in shallow permeable sediments from the northeastern Gulf of Mexico. Estuar Coast Shelf Sci 84: 202–208.
Arnosti C, Steen AD, Ziervogel K, Ghobrial S, Jeffrey WH . (2011). Latitudinal gradients in degradation of marine dissolved organic carbon. PLoS One 6: e28900.
Arnosti C, Fuchs BM, Amann R, Passow U . (2012). Contrasting extracellular enzyme activities of particle associated bacteria from distinct provinces of the North Atlantic Ocean. Front Microbiol 3: 425.
Azam F . (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280: 694–695.
Azam F, Fuhrman JA . (1984). Measurement of bacterioplankton growth in the sea and its regulation by environmental conditions. In: Hobbie JE, Williams PJ, (eds) Heterotrophic Activity in the Sea. Plenum Publishing Corp: New York, pp 179–196.
Azúa I, Unanue M, Ayo B, Artolozaga I, Arrieta JM, Iriberri J . (2003). Influence of organic matter quality in the cleavage of polymers by marine bacterial communities. J Plankton Res 25: 1451–1460.
Baltar F, Arístegui J, Sintes E, Van Aken HM, Gasol JM, Herndl GJ . (2009). Prokaryotic extracellular enzymatic activity in relation to biomass production and respiration in the meso-and bathypelagic waters of the (sub) tropical Atlantic. Environ Microbiol 11: 1998–2014.
Baltar F, Arístegui J, Gasol JM, Sintes E, Van Aken HM, Herndl GJ . (2010). High dissolved extracellular enzymatic activity in the deep central Atlantic Ocean. Aquat Microb Ecol 58: 287–302.
Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E et al. (2006). Whole genome analysis of the marine Bacteroidetes “Gramella forsetii” reveals adaptations to degradation of polymeric organic matter. Environ Microbiol 8: 2201–2213.
Becquevort S, Rousseau V, Lancelot C . (1998). Major and comparable roles for free-living and attached bacteria in the degradation of Phaeocystis-derived organic matter in Belgian coastal waters of the North Sea. Aquat Microb Ecol 14: 39–48.
Benner R, Pakulski J, McCarthy M, Hedges JI, Hatcher PG . (1992). Bulk chemical characteristics of dissolved organic matter in the ocean. Science 255: 1561.
Biersmith A, Benner R . (1998). Carbohydrates in phytoplankton and freshly produced dissolved organic matter. Mar Chem 63: 131–144.
Brettar I, Christen R, Hofle MG . (2012). Analysis of bacterial core communities in the central Baltic by comparative RNA-DNA-based fingerprinting provides links to structure-function relationships. ISME J 6: 195–212.
Burke C, Steinberg P, Rusch DB, Kjelleberg S, Thomas T . (2011). Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA 108: 14288–14293.
Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K . (2008). Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3: 283–295.
Cléroux C, Lynch-Stieglitz J, Schmidt MW, Cortijo E, Duplessy J-C . (2009). Evidence for calcification depth change of Globorotalia truncatulinoides between deglaciation and Holocene in the Western Atlantic Ocean. Mar Micropaleontol 73: 57–61.
Crump BC, Baross JA, Simenstad CA . (1998). Dominance of particle-attached bacteria in the Columbia River estuary, USA. Aquat Microb Ecol 14: 7–18.
Crump B, Armbrust E, Baross J . (1999). Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol 65: 3192–3204.
Crump BC, Hopkinson CS, Sogin ML, Hobbie JE . (2004). Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol 70: 1494–1505.
DeLong EF, Franks DG, Alldredge AL . (1993). Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr 38: 924–934.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al. (2006). Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311: 496–503.
Dempster EL, Pryor KV, Francis D, Young JE, Rogers HJ . (1999). Rapid DNA extraction from ferns for PCR-based techniques. Biotechniques 27: 66–68.
Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA 105: 7774–7778.
Garneau MČ, Vincent WF, Terrado R, Lovejoy C . (2009). Importance of particle-associated bacterial heterotrophy in a coastal Arctic ecosystem. J Marine Syst 75: 185–197.
Ghiglione JF, Conan P, Pujo-Pay M . (2009). Diversity of total and active free-living vs particle-attached bacteria in the euphotic zone of the NW Mediterranean Sea. FEMS Microbiol Lett 299: 9–21.
Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W et al. (2003). Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA 100: 8298.
Grossart H-P . (2010). Ecological consequences of bacterioplankton lifestyles: changes in concepts are needed. Environ Microb Reports 2: 706–714.
Grossart H-P, Engel A, Arnosti C, De La Rocha C, Murray A, Passow U . (2007). Microbial dynamics in autotrophic and heterotrophic seawater mesocosms: III Organic matter fluxes. Aquat Microb Ecol 49: 143–156.
Guindon S, Gascuel O . (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704.
Hamady M, Lozupone C, Knight R . (2009). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27.
Haynes K, Hofmann TA, Smith CJ, Ball AS, Underwood GJC, Osborn AM . (2007). Diatom-derived carbohydrates as factors affecting bacterial community composition in estuarine sediments. Appl Environ Microbiol 73: 6112–6124.
Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N et al. (2010). Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330: 204–208.
Hedges JI . (1992). Global biogeochemical cycles: progress and problems. Mar Chem 39: 67–93.
Herndl GJ . (1988). Ecology of amorphous aggregations (marine snow) in the Northern Adriatic Sea. II. Microbial density and activity in marine snow and its implication to overall pelagic processes. Mar Ecol Prog Ser 48: 265–275.
Holm-Hansen O, Sutcliffe WH Jr, Sharp J . (1968). Measurement of deoxyribonucleic acid in the ocean and its ecological significance. Limnol Oceanogr 13: 507–514.
Huber T, Faulkner G, Hugenholtz P . (2004). Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20: 2317–2319.
Iriberri J, Unanue M, Ayo B, Barcina I, Egea L . (1990). Bacterial production and growth rate estimation from [3H]thymidine incorporation for attached and free-living bacteria in aquatic systems. Appl Environ Microbiol 56: 483–487.
Kameo K, Shearer MC, Droxler AW, Mita I, Watanabe R, Sato T . (2004). Glacial-interglacial surface water variations in the Caribbean Sea during the last 300 ky based on calcareous nannofossil analysis. Palaeogeogr Palaeoclimatol Palaeoecol 212: 65–76.
Karner M, Herndl G . (1992). Extracellular enzymatic activity and secondary production in free-living and marine snow-associated bacteria. Marine Biol 113: 341–347.
La Cono V, Tamburini C, Genovese L, Spada GL, Denaro R, Yakimov MM . (2009). Cultivation-independent assessment of the bathypelagic archaeal diversity of Tyrrhenian Sea: Comparative study of rDNA and rRNA-derived libraries and influence of sample decompression. Deep Sea Res II 56: 768–773.
Lauro FM, Bartlett DH . (2008). Prokaryotic lifestyles in deep sea habitats. Extremophiles 12: 15–25.
Lee C, Wakeham SG, Arnosti C . (2004). Particulate organic matter in the sea: the composition conundrum. Ambio 33: 559–568.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucl Acids Res 32: 1363–1371.
Martinez J, Smith D, Steward G, Azam F . (1996). Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat Microb Ecol 10: 223–230.
Marshall KT, Morris RM . (2012). Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J 7: 452–455.
Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL et al. (2012). Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J 6: 1715–1727.
Mills HJ, Martinez RJ, Story S, Sobecky PA . (2005). Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl Environ Microbiol 71: 3235–3247.
Moeseneder M, Winter C, Herndl G . (2001). Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol Oceanogr 46: 95–107.
Moeseneder MM, Arrieta JM, Herndl GJ . (2005). A comparison of DNA-and RNA-based clone libraries from the same marine bacterioplankton community. FEMS Microbiol Ecol 51: 341–352.
Mopper K, Zhou J, Sri Ramana K, Passow U, Dam HG, Drapeau DT . (1995). The role of surface-active carbohydrates in the flocculation of a diatom bloom in a mesocosm. Deep Sea Res II 42: 47–73.
Murray AE, Arnosti C, De La Rocha CL, Grossart HP, Passow U . (2007). Microbial dynamics in autotrophic and heterotrophic seawater mesocosms. II. Bacterioplankton community structure and hydrolytic enzyme activities. Aquat Microb Ecol 49: 123–141.
O’Connor BM, Fine RA, Olson DB . (2005). A global comparison of subtropical underwater formation rates. Deep Sea Res I 52: 1569–1590.
Painter TJ Algal polysaccharides. (1983). In: Aspinall GO, (ed) The Polysaccharides vol. 2. Academic Press: New York, pp 195–285.
Pantoja S, Lee C . (1999). Peptide decomposition by extracellular hydrolysis in coastal seawater and salt marsh sediment. Mar Chem 63: 273–291.
Pommier T, Canbäck B, Riemann L, Boström KH, Simu K, Lundberg P et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880.
Porter KG, Feig YS . (1980). The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25: 943–948.
Posada D . (2008). jModelTest: Phylogenetic Model Averaging. Mol Biol Evol 25: 1253–1256.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res 35: 7188–7196.
R Core Team. (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, http://www.R-project.org/.
Rappé MS, Kemp PF, Giovannoni SJ . (1997). Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr 42: 811–826.
Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA et al. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047.
Schattenhofer M, Fuchs BM, Amann R, Zubkov MV, Tarran GA, Pernthaler J . (2009). Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ Microbiol 11: 2078–2093.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Selje N, Simon M . (2003). Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquat Microb Ecol 30: 221–237.
Simon M, Alldredge AL, Azam F . (1990). Bacterial carbon dynamics on marine snow. Mar Ecol Prog Ser 65: 205–211.
Smith DC, Simon M, Alldredge A, Azam F . (1992). Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359: 139–142.
Smith MW, Allen LZ, Allen AE, Herfort L, Simon HM . (2013). Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front Microbiol 4: 120.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA 103: 12115–12120.
Somville M, Billen G . (1983). A method for determining exoproteolytic activity in natural waters. Limnol Oceangr 28: 109–193.
Steen AD, Hamdan LJ, Arnosti C . (2008). Dynamics of dissolved carbohydrates in the Chesapeake Bay: insights from enzyme activities, concentrations, and microbial metabolism. Limnol Oceanogr 53: 936–947.
Steen AD, Ziervogel K, Ghobrial S, Arnosti C . (2012). Functional variation among polysaccharide-hydrolyzing microbial communities in the Gulf of Mexico. Marine Chem 138–139: 13–20.
Stewart F, Dalsgaard T, Young CR, Thamdrup B, Revsbech NP, Ulloa O, Canfield DE, DeLong EF . (2012). Experimental incubations elicit profound changes in community transcription in OMZ bacterioplankton. PLoS One 7 (5): e37118.
Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300.
Suzuki R, Shimodaira H . (2011). pvclust: Hierarchical Clustering with P-Values via Multiscale Bootstrap Resampling. R package version 1.2-2. http://CRAN.R-project.org/package=pvclust.
Swofford DL . (2003) PAUP*. Phylogenetic Analysis Using Parsimony and Other Methods. Version 4. Sinauer Associates: Sunderland, MA.
Teske A, Durbin A, Ziervogel K, Cox C, Arnosti C . (2011). Microbial community composition and function in permanently cold seawater and sediments from an arctic fjord of Svalbard. Appl Environ Microbiol 77: 208–218.
Teske A, Hinrichs KU, Edgcomb V, de Vera Gomez A, Kysela D, Sylva SP et al. (2002). Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl Environ Microbiol 68: 1994–2008.
Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611.
Thompson JR, Marcelino LA, Polz MF . (2002). Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res 30: 2083–2088.
Vetter Y, Deming J . (1999). Growth rates of marine bacterial isolates on particulate organic substrates solubilized by freely released extracellular enzymes. Microb Ecol 37: 86–94.
Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG et al. (2009). Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326: 578–582.
Wegner C-E, Richter-Heitmann T, Klindworth A, Klockow C, Richter M, Achstetter T et al. (2013). Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar Genomics 9: 51–61.
Weiner RM, Taylor LE, Henrissat B, Hauser L, Land M, Coutinho PM et al. (2008). Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans Strain 2-40T. PLoS Genet 4: e1000087.
Weiss MS, Abele U, Weckesser J, Welte W, Schiltz E, Schulz GE . (1991). Molecular architecture and electrostatic properties of a bacterial porin. Science 254: 1627–1630.
Wietz M, Gram L, Jorgensen B, Schramm A . (2010). Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat Microb Ecol 61: 179–189.
Worthington LV . (1982). North Atlantic circulation and water mass formation. J Mar Res 40: xii–xxii.
Worthington LV . (1976) On the North Atlantic circulation. John Hopkins Oceanographic Studies vol. VI. The John Hopkins University Press: Baltimore and London, p 110.
Wright T, Vergin K, Boyd P, Giovannoni S . (1997). A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol 63: 1441–1448.
Yoshinaga I, Fukami K, Ishida Y . (1991). Comparison of DNA and protein synthesis rates of bacterial assemblages between coral reef waters and pelagic waters in tropical ocean. Mar Ecol Prog Ser 76: 167–174.
Zhang R, Liu B, Lau SCK, Ki J-S, Qian P-Y . (2007). Particle-attached and free-living bacterial communities in a contrasting marine environment: Victoria Harbor, Hong Kong. FEMS Microbiol Ecol 61: 496–508.
Ziervogel K, Arnosti C . (2008). Polysaccharide hydrolysis in aggregates and free enzyme activity in aggregate-free seawater from the northeastern Gulf of Mexico. Environ Microbiol 10: 289–299.
Ziervogel K, Steen AD, Arnosti C . (2010). Changes in the spectrum and rates of extracellular enzyme activities in seawater following aggregate formation. Biogeosciences 7: 1007–1015.
Zimmerman AE, Martiny AC, Allison SD . (2013). Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J 13: 1–13.
Zoppini A, Puddu A, Fazi S, Rosati M, Sist P . (2005). Extracellular enzyme activity and dynamics of bacterial community in mucilaginous aggregates of the northern Adriatic Sea. Sci Total Environ 353: 270–286.
Acknowledgements
Patrick Gibson and the captain and crew of the R/V Cape Hatteras kindly collected the offshore waters for us; Dr Steven Ross, chief scientist, gave permission to collect samples. We thank Sherif Ghobrial for assistance with sample processing in the lab. This project was supported by NSF (OCE-0848703 and -1332881 to CA). We are very grateful to Antje Boetius and two anonymous reviewers for their suggestions that considerably improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
D'Ambrosio, L., Ziervogel, K., MacGregor, B. et al. Composition and enzymatic function of particle-associated and free-living bacteria: a coastal/offshore comparison. ISME J 8, 2167–2179 (2014). https://doi.org/10.1038/ismej.2014.67
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.67
Keywords
This article is cited by
-
Carbohydrates and carbohydrate degradation gene abundance and transcription in Atlantic waters of the Arctic
ISME Communications (2023)
-
Reduced bacterial mortality and enhanced viral productivity during sinking in the ocean
The ISME Journal (2022)
-
Ectohydrolytic enzyme activities of bacteria associated with Orbicella annularis coral
Coral Reefs (2021)
-
Marine Group-II archaea dominate particle-attached as well as free-living archaeal assemblages in the surface waters of Kongsfjorden, Svalbard, Arctic Ocean
Antonie van Leeuwenhoek (2021)
-
Changes in salinity and temperature drive marine bacterial communities’ structure at Potter Cove, Antarctica
Polar Biology (2019)