Abstract
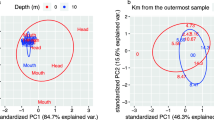
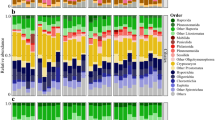
Microbial-driven organic matter (OM) degradation is a cornerstone of benthic community functioning, but little is known about the relation between OM and community composition. Here we use Rhône prodelta sediments to test the hypothesis that OM quality and source are fundamental structuring factors for bacterial communities in benthic environments. Sampling was performed on four occasions corresponding to contrasting river-flow regimes, and bacterial communities from seven different depths were analyzed by pyrosequencing of 16S rRNA gene amplicons. The sediment matrix was characterized using over 20 environmental variables including bulk parameters (for example, total nitrogen, carbon, OM, porosity and particle size), as well as parameters describing the OM quality and source (for example, pigments, total lipids and amino acids and δ13C), and molecular-level biomarkers like fatty acids. Our results show that the variance of the microbial community was best explained by δ13C values, indicative of the OM source, and the proportion of saturated or polyunsaturated fatty acids, describing OM lability. These parameters were traced back to seasonal differences in the river flow, delivering OM of different quality and origin, and were directly associated with several frequent bacterial operational taxonomic units. However, the contextual parameters, which explained at most 17% of the variance, were not always the key for understanding the community assembly. Co-occurrence and phylogenetic diversity analysis indicated that bacteria–bacteria interactions were also significant. In conclusion, the drivers structuring the microbial community changed with time but remain closely linked with the river OM input.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Achenbach LA, Michaelidou U, Bruce RA, Fryman J, Coates JD . (2001). Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int J Syst Evol Microbiol 51: 527–533.
Aller JY, Aller RC . (2004). Physical disturbance creates bacterial dominance of benthic biological communities in tropical deltaic environments of the Gulf of Papua. Cont Shelf Res 24: 2395–2416.
Aller JY, Aller RC, Kemp PF, Chistoserdov AY, Madrid VM . (2010). Fluidized muds: a novel setting for the generation of biosphere diversity through geologic time. Geobiology 8: 169–178.
Aller RC . (1998). Mobile deltaic and continental shelf muds as suboxic, fluidized bed reactors. Mar Chem 61: 143–155.
Aloisi JC, Millot C, Monaco A, Pauc H . (1979). Suspensate dynamics and mechanisms of sediment genesis on the Gulf of Lion continental-shelf. C R Acad Sci Hebd Seances Acad Sci D289: 879–882.
Antonelli C, Eyrolle F, Rolland B, Provansal M, Sabatier F . (2008). Suspended sediment and 137Cs fluxes during the exceptional December 2003 flood in the Rhône River, southeast France. Geomorphology 95: 350–360.
Baldock JA, Masiello CA, Gélinas Y, Hedges JI . (2004). Cycling and composition of organic matter in terrestrial and marine ecosystems. Mar Chem 92: 39–64.
Barberan A, Bates ST, Casamayor EO, Fierer N . (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6: 343–351.
Bianchi TS . (2011). The role of terrestrially derived organic carbon in the coastal ocean: A changing paradigm and the priming effect. Proc Natl Acad Sci USA 108: 19473–19481.
Bianchi TS, Canuel EA . (2011). Lipids: fatty acids. In: Chemical Biomarkers in Aquatic Ecosystems. Princeton University Press.
Bienhold C, Boetius A, Ramette A . (2012). The energy-diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J 6: 724–732.
Blanquer A, Uriz MJ, Galand PE . (2013). Removing environmental sources of variation to gain insight on symbionts vs. transient microbes in high and low microbial abundance sponges. Environ Microbiol 15: 3008–3019.
Bourgeois S, Pruski AM, Sun M-Y, Buscail R, Lantoine F, Kerhervé P et al. (2011). Distribution and lability of land-derived organic matter in the surface sediments of the Rhône prodelta and the adjacent shelf (Mediterranean Sea, France): a multi proxy study. Biogeosciences 8: 3107–3125.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Cathalot C, Rabouille C, Pastor L, Deflandre B, Viollier E, Buscail R et al. (2010). Temporal variability of carbon recycling in coastal sediments influenced by rivers: assessing the impact of flood inputs in the Rhône River prodelta. Biogeosciences 7: 1187–1205.
Cathalot C, Rabouille C, Tisnérat-Laborde N, Toussaint F, Kerhervé P, Buscail R et al. (2013). The fate of river organic carbon in coastal areas: a study in the Rhône River delta using multiple isotopic (δ13C, Δ14C) and organic tracers. Geochim Cosmochim Acta 118: 33–55.
Cauwet G, Gadel F, de Souza Sierra MM, Donard O, Ewald M . (1990). Contribution of the Rhône River to organic carbon inputs to the northwestern Mediterranean Sea. Cont Shelf Res 10: 1025–1037.
Charles F, Guarini-Coston J, Lantoine F, Guarini JM, Yücel M . (2014). Ecogeochemical fate of coarse organic particles in sediments of the Rhône River prodelta. Estuar Coast Shelf Sci 141: 97–103.
Darnaude A, Salen-Picard C, Polunin NC, Harmelin-Vivien M . (2004). Trophodynamic linkage between river runoff and coastal fishery yield elucidated by stable isotope data in the Gulf of Lions (NW Mediterranean). Oecologia 138: 325–332.
Dauwe B, Middelburg JJ . (1998). Amino acids and hexosamines as indicators of organic matter degradation state in North Sea sediments. Limnol Oceanogr 43: 782–798.
Diamond JM . (1975). Assembly of species communities. In: Cody ML, Diamond JM (eds) Ecology and Evolution of Communities. Belknap Press: Cambridge, MA, pp 342–444.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Efron B . (2004). Large-scale simultaneous hypothesis testing: the choice of a null hypothesis. J Am Stat Assoc 99: 96–104.
Fagervold SK, Bessette S, Romano C, Martin D, Plyuscheva M, LeBris N et al. (2013). Microbial communities associated with the degradation of oak wood in the Blanes submarine canyon and its adjacent open slope (NW Mediterranean). Prog Oceanogr 118: 137–143.
Finneran KT, Johnsen CV, Lovley DR . (2003). Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int J Syst Evol Microbiol 53: 669–673.
Franco MA, De Mesel I, Diallo MD, Van der Gucht K, Van Gansbeke D, Van Rijswijk P et al. (2007). Effect of phytoplankton bloom deposition on benthic bacterial communities in two contrasting sediments in the southern North Sea. Aquat Microb Ecol 48: 241–254.
Froelich P, Klinkhammer G, Bender M, Luedtke N, Heath G, Cullen D et al. (1979). Early oxidation of organic matter in pelagic sediments of the eastern Atlantic: suboxic diagenesis. Geochim Cosmochim Acta 43: 1075–1090.
Gobet A, Boeer SI, Huse SM, van Beusekom JEE, Quince C, Sogin ML et al. (2012). Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J 6: 542–553.
Gotelli NJ et al. (1997). Co-occurrence of Australian land birds: Diamond's assembly rules revisited. Oikos 80: 311–324.
Harmelin-Vivien M, Dierking J, Banaru D, Fontaine MF, Arlhac D . (2010). Seasonal variation in stable C and N isotope ratios of the Rhône River inputs to the Mediterranean Sea (2004-2005). Biogeochemistry 100: 139–150.
Harmelin-Vivien M, Loizeau V, Mellon C, Beker B, Arlhac D, Bodiguel X et al. (2008). Comparison of C and N stable isotope ratios between surface particulate organic matter and microphytoplankton in the Gulf of Lions (NW Mediterranean). Cont Shelf Res 28: 1911–1919.
Hedges JI, Kiel RG, Benner R . (1997). What happens to terrestrial organic matter in the ocean? Org Geochem 27: 195–212.
Higueras M, Kerhervé P, Sanchez-Vidal A, Calafat A, Ludwig W, Verdoit-Jarraya M et al. (2014). Biogeochemical characterization of the riverine organic matter transferred to the NW Mediterranean Sea. Biogeosciences 11: 157–172.
Horner-Devine MC, Bohannan BJM . (2006). Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87: S100–S108.
Horner-Devine MC, Silver JM, Leibold MA, Bohannan BJM, Colwell RK, Fuhrman JA et al. (2007). A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88: 1345–1353.
Hu JF, Zhang HB, Peng PA . (2006). Fatty acid composition of surface sediments in the subtropical Pearl River estuary and adjacent shelf, Southern China. Estuar Coast Shelf Sci 66: 346–356.
Jorgensen SL, Hannisdal B, Lanzen A, Baumberger T, Flesland K, Fonseca R et al. (2012). Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci USA 109: E2846–E2855.
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464.
Kerhervé P, Minagawa M, Heussner S, Monaco A . (2001). Stable isotopes (C-13/C-12 and N-15/N-14) in settling organic matter of the northwestern Mediterranean Sea: biogeochemical implications. Oceanologica Acta 24: S77–S85.
Kim J-H, Zarzycka B, Buscail R, Peterse F, Bonnin J, Ludwig W et al. (2010). Contribution of river-borne soil organic carbon to the Gulf of Lions (NW Mediterranean). Limnol Oceanogr 55: 507–518.
Koeppel AF, Wu M . (2014). Species matter: the role of competition in the assembly of congeneric bacteria. ISME J 8: 531–540.
Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R . (2010). Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Meth 7: 813–819.
Lansard B, Rabouille C, Denis L, Grenz C . (2009). Benthic remineralization at the land-ocean interface: A case study of the Rhône River (NW Mediterranean Sea). Estuar Coast Shelf Sci 81: 544–554.
Lansard B, Rabouille C, Denis L, Grenz C . (2008). In situ oxygen uptake rates by coastal sediments under the influence of the Rhône River (NW Mediterranean Sea). Cont Shelf Res 28: 1501–1510.
Leschine S, Paster B, Canale-Parola E . (2006). Free-living saccharolytic Spirochetes: the genus Spirochaeta. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The Prokaryotes. Springer: New York, pp 195–210.
Lindroth P, Mopper K . (1979). High performance liquid chromatographic determination of subpicomole amounts of amino acids by precolumn fluorescence derivatization with ortho-phthaldialdehyde. Anal Chem 51: 1667–1674.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Lozupone CA, Hamady M, Kelley ST, Knight R . (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73: 1576–1585.
Madrid VM, Aller JY, Aller RC, Chistoserdov AY . (2001). High prokaryote diversity and analysis of community structure in mobile mud deposits off French Guiana: identification of two new bacterial candidate divisions. FEMS Microbiol Ecol 37: 197–209.
Madrid VM, Aller RC, Aller JY, Chistoserdov AY . (2006). Evidence of the activity of dissimilatory sulfate-reducing prokaryotes in nonsulfidogenic tropical mobile muds. FEMS Microbiol Ecol 57: 169–181.
Marion C, Dufois F, Arnaud M, Vella C . (2010). In situ record of sedimentary processes near the Rhône River mouth during winter events (Gulf of Lions, Mediterranean Sea). Cont Shelf Res 30: 1095–1107.
McInerney MJ, Sieber JR, Gunsalus RP . (2009). Syntrophy in anaerobic global carbon cycles. Curr Opin Biotech 20: 623–632.
Miralles J, Arnaud M, Radakovitch O, Marion C, Cagnat X . (2006). Radionuclide deposition in the Rhône River Prodelta (NW Mediterranean sea) in response to the December 2003 extreme flood. Mar Geol 234: 179–189.
Neveux J, Lantoine F . (1993). Spectrofluorometric assay of chlorophylls and pheopigments using the least squares approximation technique. Deep Sea Res Part 1 Oceanogr Res Pap 40: 1747–1765.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara B et al. (2012) vegan: Community ecology packagehttp://cran.r-project.org/web/packages/vegan/index.html.
Paster BJ, Dewhirst FE, Weisburg WG, Tordoff LA, Fraser GJ, Hespell RB et al. (1991). Phylogenetic analysis of the spirochetes. J Bacteriol 173: 6101–6109.
Pastor L, Cathalot C, Deflandre B, Viollier E, Soetaert K, Meysman FJR et al. (2011). Modeling biogeochemical processes in sediments from the Rhône River prodelta area (NW Mediterranean Sea). Biogeosciences 8: 1351–1366.
Polymenakou PN, Bertilsson S, Tselepides A, Stephanou EG . (2005). Links between geographic location, environmental factors, and microbial community composition in sediments of the Eastern Mediterranean Sea. Microb Ecol 49: 367–378.
Pont D . (1997). Les débits solides du Rhône à proximité de son embouchure: données récentes (1994-1995). Rev Geogr Lyon 72: 23–33.
Pontarp M, Canbäck B, Tunlid A, Lundberg P . (2012). Phylogenetic analysis suggests that habitat filtering is structuring marine bacterial communities across the globe. Microb Ecol 64: 8–17.
Price MN, Dehal PS, Arkin AP . (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596.
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ . (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics, 12.
Radakovitch O, Charmasson S, Arnaud M, Bouisset P . (1999). Pb-210 and caesium accumulation in the Rhône delta sediments. Estuar Coast Shelf Sci 48: 77–92.
Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ et al. (2011). Detecting novel associations in large data sets. Science 334: 1518–1524.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Sempéré R, Charrière B, Van Wambeke F, Cauwet G . (2000). Carbon inputs of the Rhône River to the Mediterranean Sea: biogeochemical implications. Global Biogeochem Cycles 14: 669–681.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2503.
Stegen JC, Lin X, Konopka AE, Fredrickson JK . (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J 6: 1653–1664.
Stone L, Roberts A . (1990). The checkerboard score and species distributions. Oecologia 85: 74–79.
Teske A, Alm E, Regan JM, Toze S, Rittmann BE, Stahl DA . (1994). Evolutionary relationships among ammonia-oxidizing and nitrite-oxidizing bacteria. J Bacteriol 176: 6623–6630.
Vandieken V, Thamdrup B . (2013). Identification of acetate-oxidizing bacteria in a coastal marine surface sediment by RNA-stable isotope probing in anoxic slurries and intact cores. FEMS Microbiol Ecol 84: 373–386.
Vergin KL, Urbach E, Stein JL, DeLong EF, Lanoil BD, Giovannoni SJ . (1998). Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl Environ Microbiol 64: 3075–3078.
Volkman JK . (2006). Lipid markers for marine organic matter. In Volkman JK (ed) Marine Organic Matter: Biomarkers, Isotopes and DNA, The Handbook of Environmental Chemistry. Springer: Berlin, Heidelberg, pp 27–70.
Voβ M, Struck U . (1997). Stable nitrogen and carbon isotopes as indicator of eutrophication of the Oder river (Baltic sea). Mar Chem 59: 35–49.
Wang J, Soininen J, He J, Shen J . (2012). Phylogenetic clustering increases with elevation for microbes. Environ Micro Rep 4: 217–226.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Webb CO, Ackerly DD, Kembel SW . (2008). Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24: 2098–2100.
Webb CO, Ackerly DD, McPeek MA, Donoghue MJ . (2002). Phylogenies and community ecology. Annu Rev Ecol Syst 33: 475–505.
Willems A, Busse J, Goor M, Pot B, Falsen E, Jantzen E et al. (1989). Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (Formerly Pseudomonas flava), Hydrogenophaga palleronii (Formerly Pseudomonas palleronii), Hydrogenophaga pseudoflava (Formerly Pseudomonas pseudoflava and ‘Pseudomonas carboxydoflava’), and Hydrogenophaga taeniospiralis (Formerly Pseudomonas taeniospiralis). Int J Syst Bacteriol 39: 319–333.
Wolin MJ, Miller TL . (1987). Bioconversion of organic carbon to CH4 and CO2. Geomicrobiol J 5: 239–259.
Acknowledgements
We thank Christophe Rabouille, chief scientist of the MESURHOBENT cruises and coordinator of the MERMEX-RIVERS program. We thank the captain and crew of the RV Tethys II, as well as numerous colleagues, for their hard work at sea and in the laboratory. In particular, we thank Béatrice Rivière and Lionel Feuillassier from LECOB for their help with pigment and grain size analyses. Pigment concentration calculations were made using the calibration developed by François Lantoine (LECOB). We are grateful to Matthew Oliver (University of Delaware, USA) for help with R scripts for the MIC analysis, to Jeff Ghiglione and Dimitri Kalenitchenko for help with statistical analysis and Marcelino T Suzuki for his help with installing and updating QIIME and Mothur on local computer servers. Access to molecular biology instrumentation and facilities were provided through the BIO2MAR platform (http://bio2mar.obs-banyuls.fr). This research was funded by the French program MISTRALS/MERMEX, the CNRS, UPMC and in part by the ECOPOP project led by FC in the framework of ‘EC2CO/CNRS-INSU’. SB was supported by a grant from the French Ministry of Research. PEG and SKF were supported by the Agence Nationale de la Recherche (ANR) project MICADO (ANR-11JSV7-003-01). Rhône River discharge data were provided by the server Inforhône of the “Compagnie Nationale du Rhône” (CNR). The material is original work and has not been submitted for publication elsewhere.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Fagervold, S., Bourgeois, S., Pruski, A. et al. River organic matter shapes microbial communities in the sediment of the Rhône prodelta. ISME J 8, 2327–2338 (2014). https://doi.org/10.1038/ismej.2014.86
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.86
Keywords
This article is cited by
-
Clogging modulates the copper effects on microbial communities of streambed sediments
Ecotoxicology (2023)
-
Ganga River sediments of India predominate with aerobic and chemo-heterotrophic bacteria majorly engaged in the degradation of xenobiotic compounds
Environmental Science and Pollution Research (2023)
-
Does hydrological connectivity control functional characteristics of artificial wetland communities? Evidence from the Rhône River
Aquatic Sciences (2022)
-
Phytoplankton settling quality has a subtle but significant effect on sediment microeukaryotic and bacterial communities
Scientific Reports (2021)
-
Fluoride contributes to the shaping of microbial community in high fluoride groundwater in Qiji County, Yuncheng City, China
Scientific Reports (2019)