Abstract
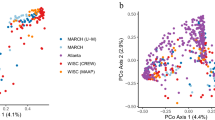
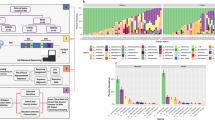
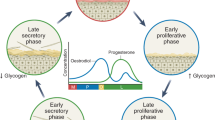
Bacterial communities colonizing the reproductive tracts of primates (including humans) impact the health, survival and fitness of the host, and thereby the evolution of the host species. Despite their importance, we currently have a poor understanding of primate microbiomes. The composition and structure of microbial communities vary considerably depending on the host and environmental factors. We conducted comparative analyses of the primate vaginal microbiome using pyrosequencing of the 16S rRNA genes of a phylogenetically broad range of primates to test for factors affecting the diversity of primate vaginal ecosystems. The nine primate species included: humans (Homo sapiens), yellow baboons (Papio cynocephalus), olive baboons (Papio anubis), lemurs (Propithecus diadema), howler monkeys (Alouatta pigra), red colobus (Piliocolobus rufomitratus), vervets (Chlorocebus aethiops), mangabeys (Cercocebus atys) and chimpanzees (Pan troglodytes). Our results indicated that all primates exhibited host-specific vaginal microbiota and that humans were distinct from other primates in both microbiome composition and diversity. In contrast to the gut microbiome, the vaginal microbiome showed limited congruence with host phylogeny, and neither captivity nor diet elicited substantial effects on the vaginal microbiomes of primates. Permutational multivariate analysis of variance and Wilcoxon tests revealed correlations among vaginal microbiota and host species-specific socioecological factors, particularly related to sexuality, including: female promiscuity, baculum length, gestation time, mating group size and neonatal birth weight. The proportion of unclassified taxa observed in nonhuman primate samples increased with phylogenetic distance from humans, indicative of the existence of previously unrecognized microbial taxa. These findings contribute to our understanding of host–microbe variation and coevolution, microbial biogeography, and disease risk, and have important implications for the use of animal models in studies of human sexual and reproductive diseases.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A et al. (2013). Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J 7: 1344–1353.
Amato KR . (2013). Co-evolution in context: the importance of studying gut microbiomes in wild animals. Microb Sc Med 1: 10–29 ISSN (Online) 2084-7653.
Anderson MJ . (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecology 26: 32–46.
Anderson MJ . (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62: 245–253.
Ankel-Simons F . (2007) Primate Anatomy: an Introduction 3rd edn Elsevier Academic Press: San Diego, CA, USA.
Aykroyd OE, Zuckerman S . (1938). Factors in sexual skin oedema. J Physiol 94: 13–25.
Baker GC, Smith JJ, Cowan DA . (2003). Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55: 541–555.
Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C . (2010). Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev 86 (Suppl 1): 13–15.
Boskey ER, Telsch KM, Whaley KJ, Moench TR, Cone RA . (1999). Acid production by vaginal flora in vitro is consistent with the rate and extent of vaginal acidification. Infect Immun 67: 5170–5175.
Campbell C, Fuentes A, MacKinnon K, Bearder S, Stumpf R . (2011) Primates in Perspective 2nd edn. Oxford University Press: Oxford.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
CDC. (2009). Trends in perinatal group B streptococcal disease - United States, 2000–2006. MMWR Morb Mortal Wkly Rep 58: 109–112.
Chapman T, Arnqvist G, Bangham J, Rowe L . (2003). Sexual conflict. TREE 18: 41–48.
Danielsson D, Teigen PK, Moi H . (2011). The genital econiche: focus on microbiota and bacterial vaginosis. Ann NY Acad Sci 1230: 48–48.
Davies TJ, Pedersen AB . (2008). Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc Biol Sci 275: 1695–1701.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N et al. (2010) Proc Natl Acad Sci USA 107: 11971–11975.
Dufrene M, Legendre P . (1997). Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67: 345–366.
Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ . (2011). Exploring the diversity of Gardnerella vaginalis in the genitourinary tract of monogamous couples through subtle nucleotide variation. PLoS One 6: e26732.
Fethers KA, Fairley CK, Hocking JS, Gurrin LC, Bradshaw CS . (2008). Sexual risk factors and bacterial vaginosis: a systematic review and meta-analysis. Clin Infect Dis 47: 1426–1435.
Fethers KA, Fairley CK, Morton A, Hocking JS, Hopkins C, Kennedy LJ et al. (2009). Early sexual experiences and risk factors for bacterial vaginosis. J Infect Dis 200: 1662–1670.
Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631.
Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, Jones R et al. (2007). Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol 73: 7059–7066.
Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ . (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74: 2461–2470.
Fredricks DN, Fiedler TL, Marrazzo JM . (2005). Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 353: 1899–1911.
Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X et al. (2012). Temporal dynamics of the human vaginal microbiota. Sci Transl Med 4: 132ra152.
Ganu RS, Ma J, Aagaard KM . (2013). The role of microbial communities in parturition: is there evidence of association with preterm birth and perinatal morbidity and mortality? Am J Perinatol 30: 613–624.
Gravett MG, Jin L, Pavlova SI, Tao L . (2012). Lactobacillus and Pediococcus species richness and relative abundance in the vagina of rhesus monkeys (Macaca mulatta). J Med Primatol 41: 183–190.
Harcourt AH, Harvey PH, Larson SG, Short RV . (1981). Testis weight, body weight and breeding system in primates. Nature 293: 55–57.
Hashway SA, Bergin IL, Bassis CM, Uchihashi M, Schmidt KC, Young VB et al. (2014). Impact of a hormone-releasing intrauterine system on the vaginal microbiome: a prospective baboon model. J Med Primatol 43: 89–99.
Hentschel U, Steinert M, Hacker J . (2000). Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol 8: 226–231.
Hummelen R, Fernandes AD, Macklaim JM, Dickson RJ, Changalucha J, Gloor GB et al. (2010). Deep sequencing of the vaginal microbiota of women with HIV. PLoS One 5: e12078.
Huse SM, Welch DM, Morrison HG, Sogin ML . (2010). Ironing out the wrinkles in the rare biosphere through improved OUT clustering. Environ Microbiol 12: 1889–1898.
Immerman R . (1986). Sexually transmitted disease and human evolution: survival of the ugliest? Hum Ethol Newsletter 4: 6–7.
Jaspers E, Overmann J . (2004). Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl Environ Microbiol 70: 4831–4839.
Kim TK, Thomas SM, Ho M, Sharma S, Reich CI, Frank JA et al. (2009). Heterogeneity of vaginal microbial communities within individuals. J Clin Microbiol 47: 1181–1189.
Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX et al. (1989). Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA 86: 6196–6200.
Kokko H, Ranta E, Ruxton G, Lundberg P . (2002). Sexually transmitted disease and the evolution of mating systems. Evolution 56: 1091–1100.
Leitich H, Kiss H . (2007). Asymptomatic bacterial vaginosis and intermediate flora as risk factors for adverse pregnancy outcome. Best Pract Res Clin Obstet Gynaecol 21: 375–390.
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI . (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6: 776–788.
Loehle C . (1995). Social barriers to pathogen transmission in wild animal populations. Ecology 76: 326–335.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Ma B, Forney LJ, Ravel J . (2012). Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol 66: 371–389.
MacManes MD . (2011). Promiscuity in mice is associated with increased vaginal bacterial diversity. Naturwissenschaften 98: 951–960.
Mandar R, Mikelsaar M . (1996). Transmission of mother's microflora to the newborn at birth. Biol Neonate 69: 30–35.
Marrazzo JM, Antonio M, Agnew K, Hillier SL . (2009). Distribution of genital Lactobacillus strains shared by female sex partners. J Infect Dis 199: 680–683.
Marrazzo JM, Martin DH, Watts DH, Schulte J, Sobel JD, Hillier SL et al. (2010). Bacterial vaginosis: identifying research gaps proceedings of a workshop sponsored by DHHS/NIH/NIAID. Sex Transm Dis 37: 732–744.
McGregor JA, French JI . (2000). Bacterial vaginosis in pregnancy. Obstet Gynecol Surv 55: 1–19.
Moller AP, Merino S, Brown CR, Robertson RJ . (2001). Immune defense and host sociality: a comparative study of swallows and martins. Am Nat 158: 136–145.
Moller H, Cortes D, Engholm G, Thorup J . (1998). Risk of testicular cancer with cryptorchidism and with testicular biopsy: cohort study. BMJ 317: 729.
Nunn CL, Gittleman JL, Antonovics J . (2000). Promiscuity and the primate immune system. Science 290: 1168–1170.
Nunn CL, Thrall PH, Leendertz FH, Boesch C . (2011). The spread of fecally transmitted parasites in socially-structured populations. PLoS One 6: e21677.
Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN . (2008). Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl Environ Microbiol 74: 4898–4909.
Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH et al. (2010). Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8: e1000546.
Patterson BD, Thaeler CS Jr . (1982). The mammalian baculum: hypotheses on the nature of bacular variability. J Mammal 63: 1–15.
Pedersen AB, Jones KE, Nunn CL, Altizer S . (2007). Infectious diseases and extinction risk in wild mammals. Conserv Biol 21: 1269–1279.
Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA et al. (2011). A molecular phylogeny of living primates. PLoS Genet 7: e1001342.
Pfeiler EA, Klaenhammer T . (2007). The genomics of lactic acid bacteria. Trends Microbiol 15: 546.
Preston BT, Stevenson IR, Pemberton JM, Coltman DW, Wilson K . (2003). Overt and covert competition in a promiscuous mammal: the importance of weaponry and testes size to male reproductive success. Proc Biol Sci 270: 633–640.
Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL et al. (2011). Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 1: 4680–4687.
Ravel J, Brotman RM, Gajer P, Ma B, Nandy M, Fadrosh DW et al. (2013). Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 1: 29.
Rivera AJ, Stumpf RM, Wilson B, Leigh S, Salyers AA . (2010). Baboon vaginal microbiota: an overlooked aspect of primate physiology. Am J Primatol 72: 467–474.
Rosenberg K, Trevathan W . (2002). Birth, obstetrics and human evolution. BJOG 109: 1199–1206.
Ruvolo M, Disotell TR, Allard MW, Brown WM, Honeycutt RL . (1991). Resolution of the African hominoid trichotomy by use of a mitochondrial gene sequence. Proc Natl Acad Sci USA 88: 1570–1574.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Sewankambo N, Gray RH, Wawer MJ, Paxton L, McNaim D, Wabwire-Mangen F et al. (1997). HIV-1 infection associated with abnormal vaginal flora morphology and bacterial vaginosis. Lancet 350: 546–550.
Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E . (2010). Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci USA 107: 20051–20056.
Smith RJ, Jungers WL . (1997). Body mass in comparative primatology. J Hum Evol 32: 523–559.
Spear GT, Gilbert D, Sikaroodi M, Doyle L, Green L, Gillevet PM et al. (2010). Identification of rhesus macaque genital microbiota by 16S pyrosequencing shows similarities to human bacterial vaginosis: implications for use as an animal model for HIV vaginal infection. AIDS Res Hum Retroviruses 26: 193–200.
Srinivasan S, Fredricks DN . (2008). The human vaginal bacterial biota and bacterial vaginosis. Interdiscip Perspect Infect Dis 2008: 750479.
Srinivasan S, Liu C, Mitchell CM, Fiedler TL, Thomas KK, Agnew KJ et al. (2010). Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS One 5: e10197.
Strier KB . (2007) Primate Behavioral Ecology 3rd edn Allyn and Bacon: Needham, MA, USA.
Stumpf RM, Rivera A, Yildirim S, Yeoman C, Wilson BA, Polk JD et al. (2013). The vaginal microbiome: Comparative perspectives and insights into human health and disease. Yearbook Phys Anthropol 57: 119–134.
Stumpf RM, Martinez-Mota R, Milich KM, Righini N, Shattuck MR . (2011). Sexual conflict in primates. Evol Anthropol 20: 62–75.
Tannock GW, Fuller R, Smith SL, Hall MA . (1990). Plasmid profiling of members of the family Enterobacteriaceae, lactobacilli, and bifidobacteria to study the transmission of bacteria from mother to infant. J Clin Microbiol 28: 1225–1228.
Thrall PH, Antonovics J, Bever J . (1997). Sexual transmission of disease and host mating systems: within-season reproductive success. Am Nat 149: 485–506.
Thrall PH, Antonovics J, Dobson AP . (2000). Sexually transmitted diseases in polygynous mating systems: prevalence and impact on reproductive success. Proc Biol Sci 267: 1555–1563.
van Oostrum N, De Sutter P, Meys J, Verstraelen H . (2013). Risks associated with bacterial vaginosis in infertility patients: a systematic review and meta-analysis. Hum Reprod 28: 1809–1815.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
White BA, Creedon DJ, Nelson KE, Wilson BA . (2011). The vaginal microbiome in health and disease. Trends Endocrinol Metab 22: 389–393.
Wiesenfeld HC, Hillier SL, Krohn MA, Landers DV, Sweet RL . (2003). Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin Infect Dis 36: 663–668.
Wiesenfeld HC, Hillier SL, Meyn LA, Amortegui AJ, Sweet RL . (2012). Subclinical pelvic inflammatory disease and infertility. Obstet Gynecol 120: 37–43.
Wlasiuk G, Nachman MW . (2010). Promiscuity and the rate of molecular evolution at primate immunity genes. Evolution 64: 2204–2220.
Woolhouse ME, Taylor LH, Haydon DT . (2001). Population biology of multihost pathogens. Science 292: 1109–1112.
Yeoman CJ, Thomas SM, Miller ME, Ulanov AV, Torralba M, Lucas S et al. (2013). A multi-omic systems-based approach reveals metabolic markers of bacterial vaginosis and insight into the disease. PLoS One 8: e56111.
Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL et al. (2010). Characterization of the fecal microbiome from nonhuman wild primates reveals species specific microbial communities. PLoS One 5: e13963.
Zhou X, Brotman RM, Gajer P, Abdo Z, Schuette U, Ma S et al. (2010). Recent advances in understanding the microbiology of the female reproductive tract and the causes of premature birth. Infect Dis Obstet Gynecol 2010: 737425.
Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P et al. (2007). Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J 1: 121–133.
Acknowledgements
This project was funded by UIUC RB and UIUC-Carle TR grants (to BA Wilson), NSF grant no. BCS-08-20709 (to RMS), and NSF HOMINID grant no. BCS-09-35347 (to SRL). We gratefully acknowledge Jeanne Altmann, Susan C Alberts, Jenny Tung, John Polk, and the late Abigail Salyers, as well as collaborator support from NCRR P40 grant no. RR019963 and VA contract no. VA247-P-0447 (to JDC), NIH grant no. 5R01RR016300 (to TT) grants R01RR016300 and R010D010980 to NBF, NSF grants BCS-0323553 and BCS-0323596 to SCA and JA, NSF grant BCS-0962807 to LCO, the Morris Animal Foundation support to TG, as well as the Office of the President of the Republic of Kenya, the Kenya Wildlife Service, and the Institute of Primate Research, Kenya, The Uganda Wildlife Authority, The Uganda National Council for Science and Technology, the Chimpanzee Sanctuary and Wildlife Conservation Trust, The Wildlife Conservation Society, The University of Veracruz, and Consejo Nacional de Areas Protegidas Guatemala. We also thank Michael Worobey and Anthony Yannarell for their guidance with phylogenetic and statistical analyses, respectively.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Yildirim, S., Yeoman, C., Janga, S. et al. Primate vaginal microbiomes exhibit species specificity without universal Lactobacillus dominance. ISME J 8, 2431–2444 (2014). https://doi.org/10.1038/ismej.2014.90
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.90
This article is cited by
-
The Medium Is the Message: Defining a “Normal” Vaginal Microbiome in Healthy Reproductive-Age Women
Reproductive Sciences (2023)
-
Insight into the ecology of vaginal bacteria through integrative analyses of metagenomic and metatranscriptomic data
Genome Biology (2022)
-
Age and sex-associated variation in the multi-site microbiome of an entire social group of free-ranging rhesus macaques
Microbiome (2021)
-
Interactions between reproductive biology and microbiomes in wild animal species
Animal Microbiome (2021)
-
Shifts in microbial diversity, composition, and functionality in the gut and genital microbiome during a natural SIV infection in vervet monkeys
Microbiome (2020)