Abstract
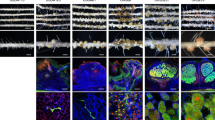
The occurrence of alternative Nod factor (NF)-independent symbiosis between legumes and rhizobia was first demonstrated in some Aeschynomene species that are nodulated by photosynthetic bradyrhizobia lacking the canonical nodABC genes. In this study, we revealed that a large diversity of non-photosynthetic bradyrhizobia, including B. elkanii, was also able to induce nodules on the NF-independent Aeschynomene species, A. indica. Using cytological analysis of the nodules and the nitrogenase enzyme activity as markers, a gradient in the symbiotic interaction between bradyrhizobial strains and A. indica could be distinguished. This ranged from strains that induced nodules that were only infected intercellularly to rhizobial strains that formed nodules in which the host cells were invaded intracellularly and that displayed a weak nitrogenase activity. In all non-photosynthetic bradyrhizobia, the type III secretion system (T3SS) appears required to trigger nodule organogenesis. In contrast, genome sequence analysis revealed that apart from a few exceptions, like the Bradyrhizobium ORS285 strain, photosynthetic bradyrhizobia strains lack a T3SS. Furthermore, analysis of the symbiotic properties of an ORS285 T3SS mutant revealed that the T3SS could have a positive or negative role for the interaction with NF-dependent Aeschynomene species, but that it is dispensable for the interaction with all NF-independent Aeschynomene species tested. Taken together, these data indicate that two NF-independent symbiotic processes are possible between legumes and rhizobia: one dependent on a T3SS and one using a so far unknown mechanism.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Aslam SN, Newman MA, Erbs G, Morrissey KL, Chinchilla D, Boller T et al. (2008). Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol 18: 1078–1083.
Bonaldi K, Gargani D, Prin Y, Fardoux J, Gully D, Nouwen N et al. (2011). Nodulation of Aeschynomene afraspera and A. indica by photosynthetic Bradyrhizobium sp. strain ORS285: the Nod-dependent versus the Nod-independent symbiotic interaction. Mol Plant Microbe Interact 24: 1359–1371.
Bonaldi K, Gourion B, Fardoux J, Hannibal L, Cartieaux F et al. (2010). Large-scale transposon mutagenesis of photosynthetic Bradyrhizobium sp. strain ORS278 reveals new genetic loci putatively important for Nod-independent symbiosis with Aeschynomene indica. Mol Plant Microbe Interact 23: 760–770.
Chaintreuil C, Arrighi JF, Giraud E, Miché L, Moulin L, Dreyfus B et al. (2013). Evolution of symbiosis in the legume genus Aeschynomene. New Phytol 200: 1247–1259.
D’Haeze W, Holsters M . (2004). Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol 12: 555–561.
Dodds PN, Rathjen JP . (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548.
Felsenstein J . (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 424–429.
Fraysse N, Couderc F, Poinsot V . (2003). Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur J Biochem 270: 1365–1380.
Gage DJ . (2002). Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol 184: 7042–7046.
Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC et al. (2007). Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316: 1307–1312.
Gourion B, Berrabah F, Ratet P, Stacey G . (2014). Rhizobium-legume symbioses: the crucial role of plant immunity. Trends Plant Sci pii: S1360–S1385 (14)00297-0.
Gueye F, Moulin L, Sylla S, Ndoye I, Béna G . (2009). Genetic diversity and distribution of Bradyrhizobium and Azorhizobium strains associated with the herb legume Zornia glochidiata sampled from across Senegal. Syst Appl Microbiol 32: 387–399.
Haag AF, Baloban M, Sani M, Kerscher B, Pierre O, Farkas A et al. (2011). Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol 9: e1001169.
Hueck CJ . (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62: 379–433.
Kelly SJ, Muszyński A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N et al. (2013). Conditional requirement for exopolysaccharide in the Mesorhizobium-Lotus symbiosis. Mol Plant Microbe Interact 26: 319–329.
Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC et al. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784.
Liang Y, Tóth K, Cao Y, Tanaka K, Espinoza C, Stacey G . (2014). Lipochitooligosaccharide recognition: an ancient story. New Phytol 204: 289–296.
Lorquin J, Molouba F, Dupuy N, Ndiaye S, Alazard D, Gillis M et al Diversity of photosynthetic Bradyrhizobium strains from stem nodules of Aeschynomene species. In Palacios R, Mora J, Newton WE (Eds). (1993) New Horizons in Nitrogen Fixation. Boston: Kluwer, 17: 683–689.
Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS et al. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 10.
Miché L, Moulin L, Chaintreuil C, Contreras-Jimenez JL, Munive-Hernández JA, Del Carmen Villegas-Hernandez M et al. (2010). Diversity analyses of Aeschynomene symbionts in Tropical Africa and Central America reveal that Nod-independent stem nodulation is not restricted to photosynthetic bradyrhizobia. Environ Microbiol 12: 2152–2164.
Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B et al. (1999). Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16 S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol 65: 3084–3094.
Montecchia MS, Kerber NL, Pucheu NL, Perticari A, García AF . (2002). Analysis of genomic diversity among photosynthetic stem-nodulating rhizobial strains from northeast Argentina. Syst Appl Microbiol 25: 423–433.
Mornico D, Miché L, Béna G, Nouwen N, Verméglio A, Vallenet D et al. (2011). Comparative genomics of Aeschynomene symbionts: insights into the ecological lifestyle of Nod-independent photosynthetic bradyrhizobia. Genes 3: 35–61.
Nagata T, Takebe L . (1970). Cell wall regeneration and cell division in isolated tobacco mesophyll protoplasts. Planta 92: 301–308.
Okazaki S, Kaneko T, Sato S, Saeki K . (2013). Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc Natl Acad Sci USA 110: 17131–17136.
Oldroyd GE, Murray JD, Poole PS, Downie JA . (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144.
Oldroyd GE . (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263.
Saitou RR, Nei M . (1987). A neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 44: 406–425.
Tampakaki AP . (2014). Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria. Front Plant Sci 5: 114.
Thompson JD, Gibson TJ, Higgins DG . (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2.3.
Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126.
Vincent JM . (1970) A Manual for the Practical Study of Root-Nodule Bacteria. Blackwell Scientific Publications: Oxford, United Kingdom.
Wang D, Yang S, Tang F, Zhu H . (2012). Symbiosis specificity in the legume: rhizobial mutualism. Cell Microbiol 14: 334–342.
Wassem R, Kobayashi H, Kambara K, Le Quéré A, Walker GC, Broughton WJ et al. (2008). TtsI regulates symbiotic genes in Rhizobium species NGR234 by binding to tts boxes. Mol Microbiol 68: 736–748.
Wilson KJ, Sessitsch A, Corbo JC, Giller KE, Akkermans AD, Jefferson RA . (1995). β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology 141: 1691–1705.
Zehner S, Schober G, Wenzel M, Lang K, Göttfert M . (2008). Expression of the Bradyrhizobium japonicum type III secretion system in legume nodules and analysis of the associated tts box promoter. Mol Plant Microbe Interact 21: 1087–1093.
Acknowledgements
This work was supported by grant from the French national research agency (ANR-BugsInaCell-13-BSV7-0013) and the Franco-Thai PHC Siam program (project SIAM N°29589XA). We thank P Mergaert (ISV, CNRS, Gif-sur-Yvette, France) for criticisms and corrections of the manuscript. We thank Tatiana Krassova-Wade (LCM, IRD, Dakar, Senegal) for providing some Bradyrhizobium strains.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Okazaki, S., Tittabutr, P., Teulet, A. et al. Rhizobium–legume symbiosis in the absence of Nod factors: two possible scenarios with or without the T3SS. ISME J 10, 64–74 (2016). https://doi.org/10.1038/ismej.2015.103
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.103
This article is cited by
-
Occurrence and diversity of stem nodulation in Aeschynomene and Sesbania legumes from wetlands of Madagascar
Scientific Reports (2024)
-
Nod–factors are dispensable for nodulation: A twist in bradyrhizobia-legume symbiosis
Symbiosis (2022)
-
Evolutionary origin and ecological implication of a unique nif island in free-living Bradyrhizobium lineages
The ISME Journal (2021)
-
Identification of type III effectors modulating the symbiotic properties of Bradyrhizobium vignae strain ORS3257 with various Vigna species
Scientific Reports (2021)
-
Rhizobia use a pathogenic-like effector to hijack leguminous nodulation signalling
Scientific Reports (2021)