Abstract
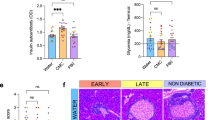
Accumulating evidence supports that the intestinal microbiome is involved in Type 1 diabetes (T1D) pathogenesis through the gut-pancreas nexus. Our aim was to determine whether the intestinal microbiota in the non-obese diabetic (NOD) mouse model played a role in T1D through the gut. To examine the effect of the intestinal microbiota on T1D onset, we manipulated gut microbes by: (1) the fecal transplantation between non-obese diabetic (NOD) and resistant (NOR) mice and (2) the oral antibiotic and probiotic treatment of NOD mice. We monitored diabetes onset, quantified CD4+T cells in the Peyer’s patches, profiled the microbiome and measured fecal short-chain fatty acids (SCFA). The gut microbiota from NOD mice harbored more pathobionts and fewer beneficial microbes in comparison with NOR mice. Fecal transplantation of NOD microbes induced insulitis in NOR hosts suggesting that the NOD microbiome is diabetogenic. Moreover, antibiotic exposure accelerated diabetes onset in NOD mice accompanied by increased T-helper type 1 (Th1) and reduced Th17 cells in the intestinal lymphoid tissues. The diabetogenic microbiome was characterized by a metagenome altered in several metabolic gene clusters. Furthermore, diabetes susceptibility correlated with reduced fecal SCFAs. In an attempt to correct the diabetogenic microbiome, we administered VLS#3 probiotics to NOD mice but found that VSL#3 colonized the intestine poorly and did not delay diabetes. We conclude that NOD mice harbor gut microbes that induce diabetes and that their diabetogenic microbiome can be amplified early in life through antibiotic exposure. Protective microbes like VSL#3 are insufficient to overcome the effects of a diabetogenic microbiome.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Akerblom HK, Knip M . (1998). Putative environmental factors in Type 1 diabetes. Diabetes Metab Rev 14: 31–67.
Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K . (2010). Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta 1801: 1175–1183.
Alam C, Bittoun E, Bhagwat D, Valkonen S, Saari A, Jaakkola U et al. (2011). Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia 54: 1398–1406.
Anderson MJ . (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
Bray JR, Curtis JT . (1957). An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol Monogr 27: 326–349.
Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S et al. (2005). Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia 48: 1565–1575.
Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray AP, Pelletier B et al. (2015). Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One 10: e0125448.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Chao A, Chazdon RL, Colwell RK, Shen TJ . (2005). A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett 8: 148–159.
de Kort S, Keszthelyi D, Masclee AA . (2011). Leaky gut and diabetes mellitus: what is the link? Obes Rev 12: 449–458.
Edgar RC . (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Ghosh S, Dai C, Brown K, Rajendiran E, Makarenko S, Baker J et al. (2011). Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status, and ion transporter gene expression. Am J Physiol Gastrointest Liver Physiol 301: G39–G49.
Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G et al. (2011). Toward defining the autoimmune microbiome for type 1 diabetes. ISME J 5: 82–91.
Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sorensen SJ et al. (2012). Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 55: 2285–2294.
Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN et al. (2012). Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol 189: 3805–3814.
Herlemann DP, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF . (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5: 1571–1579.
Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB et al. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4: 337–349.
Jaakkola I, Jalkanen S, Hanninen A . (2003). Diabetogenic T cells are primed both in pancreatic and gut-associated lymph nodes in NOD mice. Eur J Immunol 33: 3255–3264.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M . (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114.
Kim CH, Park J, Kim M . (2014). Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immune Netw 14: 277–288.
King C, Sarvetnick N . (2011). The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One 6: e17049.
Knip M . (2012). Descriptive epidemiology of type 1 diabetes-is it still in? Diabetologia 55: 1227–1230.
Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM et al. (2015). The dynamics of the human infant gut microbiome in development and in progression toward Type 1 diabetes. Cell Host Microbe 17: 260–273.
Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D . (2011). Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA 108: 11548–11553.
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16 S rRNA marker gene sequences. Nat Biotechnol 31: 814–821.
Lau K, Benitez P, Ardissone A, Wilson TD, Collins EL, Lorca G et al. (2011). Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol 186: 3538–3546.
Lee AS, Gibson DL, Zhang Y, Sham HP, Vallance BA, Dutz JP . (2010). Gut barrier disruption by an enteric bacterial pathogen accelerates insulitis in NOD mice. Diabetologia 53: 741–748.
Lee AS, Ghoreishi M, Cheng WK, Chang TY, Zhang YQ, Dutz JP . (2011). Toll-like receptor 7 stimulation promotes autoimmune diabetes in the NOD mouse. Diabetologia 54: 1407–1416.
Lee JY, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A . (2013). Contribution of the 7beta-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res 54: 3062–3069.
Macfarlane S, Macfarlane GT . (2003). Regulation of short-chain fatty acid production. Proc Nutr Soc 62: 67–72.
Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U et al. (2013). Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339: 1084–1088.
Mitrou P, Petsiou E, Papakonstantinou E, Maratou E, Lambadiari V, Dimitriadis P et al. (2015). The role of acetic acid on glucose uptake and blood flow rates in the skeletal muscle in humans with impaired glucose tolerance. Eur J Clin Nutr 69: 734–739.
Neu J, Lorca G, Kingma SD, Triplett EW . (2010). The intestinal microbiome: relationship to type 1 diabetes. Endocrinol Metab Clin North Am 39: 563–571.
Pomare EW, Branch WJ, Cummings JH . (1985). Carbohydrate fermentation in the human colon and its relation to acetate concentrations in venous blood. J Clin Invest 75: 1448–1454.
Pozzilli P, Signore A, Williams AJ, Beales PE . (1993). NOD mouse colonies around the world—recent facts and figures. Immunol Today 14: 193–196.
Prochazka M, Serreze DV, Frankel WN, Leiter EH . (1992). NOR/Lt mice: MHC-matched diabetes-resistant control strain for NOD mice. Diabetes 41: 98–106.
Puertollano E, Kolida S, Yaqoob P . (2014). Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr Opin Clin Nutr Metab Care 17: 139–144.
Rabinovitch A, Suarez-Pinzon WL, Sorensen O, Bleackley RC, Power RF . (1995). IFN-gamma gene expression in pancreatic islet-infiltrating mononuclear cells correlates with autoimmune diabetes in nonobese diabetic mice. J Immunol 154: 4874–4882.
Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM et al. (2009). Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J 3: 536–548.
Sapone A, de Magistris L, Pietzak M, Clemente MG, Tripathi A, Cucca F et al. (2006). Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes 55: 1443–1449.
Valladares R, Sankar D, Li N, Williams E, Lai KK, Abdelgeliel AS et al. (2010). Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS One 5: e10507.
Vehik K, Hamman RF, Lezotte D, Norris JM, Klingensmith G, Bloch C et al. (2007). Increasing incidence of type 1 diabetes in 0- to 17-year-old Colorado youth. Diabetes Care 30: 503–509.
Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC et al. (2008). Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455: 1109–1113.
Yanagibashi T, Hosono A, Oyama A, Tsuda M, Suzuki A, Hachimura S et al. (2013). IgA production in the large intestine is modulated by a different mechanism than in the small intestine: Bacteroides acidifaciens promotes IgA production in the large intestine by inducing germinal center formation and increasing the number of IgA+ B cells. Immunobiology 218: 645–651.
Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L et al. (2013). Gender bias in autoimmunity is influenced by microbiota. Immunity 39: 400–412.
Zhang Y, Lee AS, Shameli A, Geng X, Finegood D, Santamaria P et al. (2010). TLR9 blockade inhibits activation of diabetogenic CD8+ T cells and delays autoimmune diabetes. J Immunol 184: 5645–5653.
Acknowledgements
BAV holds the Children with Intestinal and Liver Disorders (CHILD) Foundation Chair in Pediatric Gastroenterology. BAV is also the Canada Research Chair (Tier 2) in Pediatric Gastroenterology. JPD is a senior scientist at the Child and Family Research Institute. This work was supported by operating grants to BAV from the Canadian Institutes for Health Research, to JPD and BAV from Juvenile Diabetes Research Foundation, to JPD and DLG from the Child and Family Research Institute Diabetes Catalyst Grant from the Canuck’s Foundation and to DLG from VSL#3 Pharmaceuticals and Natural Sciences and Engineering Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Brown, K., Godovannyi, A., Ma, C. et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J 10, 321–332 (2016). https://doi.org/10.1038/ismej.2015.114
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.114
This article is cited by
-
Mechanistic Insights into Immune-Microbiota Interactions and Preventive Role of Probiotics Against Autoimmune Diabetes Mellitus
Probiotics and Antimicrobial Proteins (2023)
-
Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders
Nature Reviews Gastroenterology & Hepatology (2022)
-
Inverse association between use of broad spectrum penicllin with beta-lactamase inhibitors and prevalence of type 1 diabetes mellitus in Europe
Scientific Reports (2021)
-
The gut microbiome of laboratory mice: considerations and best practices for translational research
Mammalian Genome (2021)
-
Antibiotic affects the gut microbiota composition and expression of genes related to lipid metabolism and myofiber types in skeletal muscle of piglets
BMC Veterinary Research (2020)