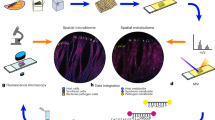
Abstract
One of the greatest challenges in microbial ecology remains to link the metabolic activity of individual cells to their taxonomic identity and localization within environmental samples. Here we combined mass-spectrometric imaging (MSI) through (matrix-assisted) laser desorption ionization time-of-flight MSI ([MA]LDI-TOF/MSI) with fluorescence in situ hybridization (FISH) to monitor antibiotic production in the defensive symbiosis between beewolf wasps and ‘Streptomyces philanthi’ bacteria. Our results reveal similar distributions of the different symbiont-produced antibiotics across the surface of beewolf cocoons, which colocalize with the producing cell populations. Whereas FISH achieves single-cell resolution, MSI is currently limited to a step size of 20–50 μm in the combined approach because of the destructive effects of high laser intensities that are associated with tighter laser beam focus at higher lateral resolution. However, on the basis of the applicability of (MA)LDI-MSI to a broad range of small molecules, its combination with FISH provides a powerful tool for studying microbial interactions in situ, and further modifications of this technique could allow for linking metabolic profiling to gene expression.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Behrens S, Loesekann T, Pett-Ridge J, Weber PK, Ng W-O, Stevenson BS et al. (2008). Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol 74: 3143–3150.
Goettler W, Kaltenpoth M, Herzner G, Strohm E . (2007). Morphology and ultrastructure of a bacteria cultivation organ: the antennal glands of female European beewolves, Philanthus triangulum (Hymenoptera, Crabronidae). Arthropod Struct Dev 36: 1–9.
Hoelscher D, Shroff R, Knop K, Gottschaldt M, Crecelius A, Schneider B et al. (2009). Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. Plant J 60: 907–918.
Kaltenpoth M, Gottler W, Herzner G, Strohm E . (2005). Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15: 475–479.
Kaltenpoth M, Goettler W, Dale C, Stubblefield JW, Herzner G, Roeser-Mueller K et al. (2006). 'Candidatus Streptomyces philanthi', an endosymbiotic streptomycete in the antennae of Philanthus digger wasps. Int J Syst Evol Microbiol 56: 1403–1411.
Kaltenpoth M, Goettler W, Koehler S, Strohm E . (2010a). Life cycle and population dynamics of a protective insect symbiont reveal severe bottlenecks during vertical transmission. Evol Ecol 24: 463–477.
Kaltenpoth M, Schmitt T, Polidori C, Koedam D, Strohm E . (2010b). Symbiotic streptomycetes in antennal glands of the South American digger wasp genus Trachypus (Hymenoptera, Crabronidae). Physiol Entomol 35: 196–200.
Kaltenpoth M, Roeser-Mueller K, Koehler S, Peterson A, Nechitaylo T, Stubblefield JW et al. (2014). Partner choice and fidelity stabilize co-evolution in a Cretaceous-age defensive symbiosis. Proc Natl Acad Sci USA 111: 6359–6364.
Kindaichi T, Ito T, Okabe S . (2004). Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl Environ Microbiol 70: 1641–1650.
Koehler S, Doubský J, Kaltenpoth M . (2013). Dynamics of symbiont-mediated antibiotic production reveal efficient long-term protection for beewolf offspring. Front Zool 10: 3.
Kroiss J, Kaltenpoth M, Schneider B, Schwinger M-G, Hertweck C, Maddula RK et al. (2010). Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol 6: 261–263.
Li T, Wu T-D, Mazeas L, Toffin L, Guerquin-Kern J-L, Leblon G et al. (2008). Simultaneous analysis of microbial identity and function using NanoSIMS. Environ Microbiol 10: 580–588.
Moraru C, Lam P, Fuchs BM, Kuypers MMM, Amann R . (2010). GeneFISH - an in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ Microbiol 12: 3057–3073.
Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F et al. (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA 105: 17861–17866.
Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF . (2001). Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293: 484–487.
Pernthaler A, Amann R . (2004). Simultaneous fluorescence in situ hybridization of mRNA and rRNA in environmental bacteria. Appl Environ Microbiol 70: 5426–5433.
Schönhuber W, Fuchs B, Juretschko S, Amann R . (1997). Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol 63: 3268–3273.
Svatoš A . (2010). Mass spectrometric imaging of small molecules. Trends Biotechnol 28: 425–434.
Svatoš A . (2011). Single-cell metabolomics comes of age: new developments in mass spectrometry profiling and imaging. Anal Chem 83: 5037–5044.
Svatoš A, Mock H-P . (2013) MALDI mass spectrometric imaging of plants. In: Weckwerth W, Kahl G (eds). The Handbook of Plant Metabolomics. Wiley-VCH: Weinheim, Germany, pp 93–110.
Yang YL, Xu YQ, Straight P, Dorrestein PC . (2009). Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol 5: 885–887.
Zwirglmaier K . (2005). Fluorescence in situ hybridisation (FISH) - the next generation. FEMS Microbiol Lett 246: 151–158.
Acknowledgements
We thank Benjamin Weiss for assistance with the FISH experiments and Taras Nechitaylo for help with the ‘S. philanthi’ cultivation. We gratefully acknowledge funding from the Max Planck Society (MK and AS) and the German Science Foundation (DFG KA2846/2-1 to MK).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Kaltenpoth, M., Strupat, K. & Svatoš, A. Linking metabolite production to taxonomic identity in environmental samples by (MA)LDI-FISH. ISME J 10, 527–531 (2016). https://doi.org/10.1038/ismej.2015.122
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.122
This article is cited by
-
Omics for deciphering oral microecology
International Journal of Oral Science (2024)
-
Visualization of metabolites and microbes at high spatial resolution using MALDI mass spectrometry imaging and in situ fluorescence labeling
Nature Protocols (2023)
-
Bacterial ectosymbionts in cuticular organs chemically protect a beetle during molting stages
The ISME Journal (2022)
-
Spatial metabolomics of in situ host–microbe interactions at the micrometre scale
Nature Microbiology (2020)
-
Die chemische Sprache von Symbiosen sichtbar machen
BIOspektrum (2020)