Abstract
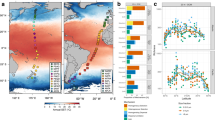
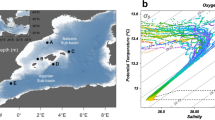
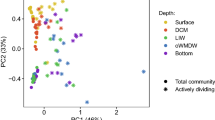
The deep-sea is the largest biome of the biosphere, and contains more than half of the whole ocean’s microbes. Uncovering their general patterns of diversity and community structure at a global scale remains a great challenge, as only fragmentary information of deep-sea microbial diversity exists based on regional-scale studies. Here we report the first globally comprehensive survey of the prokaryotic communities inhabiting the bathypelagic ocean using high-throughput sequencing of the 16S rRNA gene. This work identifies the dominant prokaryotes in the pelagic deep ocean and reveals that 50% of the operational taxonomic units (OTUs) belong to previously unknown prokaryotic taxa, most of which are rare and appear in just a few samples. We show that whereas the local richness of communities is comparable to that observed in previous regional studies, the global pool of prokaryotic taxa detected is modest (~3600 OTUs), as a high proportion of OTUs are shared among samples. The water masses appear to act as clear drivers of the geographical distribution of both particle-attached and free-living prokaryotes. In addition, we show that the deep-oceanic basins in which the bathypelagic realm is divided contain different particle-attached (but not free-living) microbial communities. The combination of the aging of the water masses and a lack of complete dispersal are identified as the main drivers for this biogeographical pattern. All together, we identify the potential of the deep ocean as a reservoir of still unknown biological diversity with a higher degree of spatial complexity than hitherto considered.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL et al. (2004). Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430: 551–554.
Agogué H, Lamy D, Neal PR, Sogin ML, Herndl GJ . (2011). Water mass-specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol Ecol 20: 258–274.
Allen LZ, Allen EE, Badger JH, McCrow JP, Paulsen IT, Elbourne LD et al. (2012). Influence of nutrients and currents on the genomic composition of microbes across an upwelling mosaic. ISME J 6: 1403–1414.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Anderson MJ . (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
Anderson MJ, Walsh DCI . (2013). PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol Monogr 83: 557–574.
Arístegui J, Gasol JM, Duarte CM, Herndl GJ . (2009). Microbial oceanography of the dark ocean’s pelagic realm. Limnol Oceanogr 54: 1501–1529.
Brown MV, Philip GK, Bunge JA, Smith MC, Bissett A, Lauro FM et al. (2009). Microbial community structure in the North Pacific ocean. ISME J 3: 1374–1386.
Brown MV, Schwalbach MS, Hewson I, Fuhrman JA . (2005). Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ Microbiol 7: 1466–1479.
Caporaso JG, Lauber CL, Walters Wa, Berg-Lyons D, Lozupone Ca, Turnbaugh PJ et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108 (Suppl): 4516–4522.
Clarke K, Ainsworth M . (1993). A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser 92: 205–219.
Cohan FM . (2002). What are bacterial species? Annu Rev Microbiol 56: 457–487.
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y et al. (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42: D633–D642.
Crump BC, Armbrust EV, Baross JA . (1999). Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol 65: 3192–3204.
DeLong EF . (1992). Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685–5689.
DeLong EF . (2003). Oceans of archaea. ASM News-American Soc Microbiol 69: 503.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U et al. (2006). Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311: 496–503.
Eloe EA, Shulse CN, Fadrosh DW, Williamson SJ, Allen EE, Bartlett DH et al. (2011). Compositional differences in particle-associated and free-living microbial assemblages from an extreme deep-ocean environment. Environ Microbiol Rep 3: 449–458.
Fox GE, Wisotzkey JD, Jurtshuk P . (1992). How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol 42: 166–170.
Fuhrman JA, Hewson I, Schwalbach MS, Steele Ja, Brown MV, Naeem S . (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109.
Galand PE, Potvin M, Casamayor EO, Lovejoy C . (2010). Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J 4: 564–576.
Ganesh S, Parris DJ, DeLong EF, Stewart FJ . (2014). Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J 8: 187–211.
Ghiglione J-F, Conan P, Pujo-Pay M . (2009). Diversity of total and active free-living vs particle-attached bacteria in the euphotic zone of the NW Mediterranean Sea. FEMS Microbiol Lett 299: 9–21.
Ghiglione JF, Galand PE, Pommier T, Pedrós-Alió C, Maas EW, Bakker K et al. (2012). Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci USA 109: 17633–17638.
Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T et al. (2009). The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139.
Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308.
Giovannoni SJ . (2012). Seasonality in ocean microbial communities. Science 335: 671–676.
Hagström Å, Pommier T, Rohwer F, Simu K, Stolte W, Svensson D et al. (2002). Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl Environ Microbiol 68: 3628–3633.
Hamdan LJ, Coffin RB, Sikaroodi M, Greinert J, Treude T, Gillevet PM . (2013). Ocean currents shape the microbiome of Arctic marine sediments. ISME J 7: 685–696.
Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH . (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10: 1–10.
Herndl GJ, Reinthaler T . (2013). Microbial control of the dark end of the biological pump. Nat Geosci 6: 718–724.
Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A et al. (2005). Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol 71: 2303–2309.
Irigoien X, Klevjer TA, Røstad A, Martinez U, Boyra G, Acuña JL et al. (2014). Large mesopelagic fishes biomass and trophic efficiency in the open ocean. Nat Commun 5: 3271.
Ivars-Martinez E, Martin-Cuadrado A-B, D’Auria G, Mira A, Ferriera S, Johnson J et al. (2008). Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J 2: 1194–1212.
Jenkins WJ . (1982). Oxygen utilization rates in North Atlantic subtropical gyre and primary production in oligotrophic systems. Nature 300: 246–248.
Karner MB, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510.
López-García P, López-López A, Moreira D, Rodríguez-Valera F . (2001). Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microbiol Ecol 36: 193–202.
López-López A, Bartual SG, Stal L, Onyshchenko O, Rodríguez-Valera F . (2005). Genetic analysis of housekeeping genes reveals a deep-sea ecotype of Alteromonas macleodii in the Mediterranean Sea. Environ Microbiol 7: 649–659.
Martín-Cuadrado A-B, López-García P, Alba J-C, Moreira D, Monticelli L, Strittmatter A et al. (2007). Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS One 2: e914.
Massana R, DeLong EF, Pedrós-Alió C . (2000). A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl Environ Microbiol 66: 1777–1787.
Massana R, Murray A, Preston C, DeLong E . (1997). Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Envir Microbiol 63: 50–56.
Minchin PR . (1987). An evaluation of the relative robustness of techniques for ecological ordination. Vegetatio 69: 89–107.
Moalic Y, Desbruyères D, Duarte CM, Rozenfeld AF, Bachraty C, Arnaud-Haond S . (2012). Biogeography revisited with network theory: retracing the history of hydrothermal vent communities. Syst Biol 61: 127–137.
Nunoura T, Takaki Y, Hirai M, Shimamura S, Makabe A, Koide O . (2015). Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc Natl Acad Sci 112: E1230–E1236.
Oden NL, Sokal RR . (1986). Directional autocorrelation: an extension of spatial correlograms to two dimensions. Syst Zool 35: 608.
Pedrós-Alió C . (2012). The rare bacterial biosphere. Ann Rev Mar Sci 4: 449–466.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Pruitt KD, Tatusova T, Brown GR, Maglott DR . (2012). NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40: D130–D135.
Quaiser A, Zivanovic Y, Moreira D, López-García P . (2011). Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5: 285–304.
R Core Team. (2014), R: A language and environment for statistical computing.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77.
Schauer R, Bienhold C, Ramette A, Harder J . (2010). Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J 4: 159–170.
Smedile F, Messina E, La Cono V, Tsoy O, Monticelli LS, Borghini M et al. (2012). Metagenomic analysis of hadopelagic microbial assemblages thriving at the deepest part of Mediterranean Sea, Matapan-Vavilov Deep. Environ Microbiol 15: 167–182.
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Stackebrandt E . (2006) Defining Taxonomic Ranks. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds). The Prokaryotes SE—3. Springer: New York, NY, USA, pp 29–57.
Stackebrandt E, Goebel BM . (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44: 846–849.
Sul WJ, Oliver Ta, Ducklow HW, Amaral-Zettler La, Sogin ML . (2013). Marine bacteria exhibit a bipolar distribution. Proc Natl Acad Sci USA pp 1–6.
Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G et al. (2015). Structure and function of the global ocean microbiome. Science 348: 1261359–1261359.
Teira E, Lebaron P, van Aken H, Herndl GJ . (2006). Distribution and activity of Bacteria and Archaea in the deep water masses of the North Atlantic. Limnol Oceanogr 51: 2131–2144.
Wang Y, Cao H, Zhang G, Bougouffa S, Lee OO, Al-Suwailem A et al. (2013). Autotrophic microbe metagenomes and metabolic pathways differentiate adjacent red sea brine pools. Sci Rep 3: 1748.
Wilkins D, van Sebille E, Rintoul SR, Lauro FM, Cavicchioli R . (2013). Advection shapes Southern Ocean microbial assemblages independent of distance and environment effects. Nat Commun 4: 2457.
Winter C, Kerros M-E, Weinbauer MG . (2009). Seasonal changes of bacterial and archaeal communities in the dark ocean: evidence from the Mediterranean Sea. Limnol Oceanogr 54: 160–170.
Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K et al. (2007). The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol 5: e16.
Zinger L, Amaral-Zettler La, Fuhrman Ja, Horner-Devine MC, Huse SM, Welch DBM et al. (2011). Global Patterns of Bacterial Beta-Diversity in Seafloor and Seawater Ecosystems. In: Gilbert, JA (ed). PLoS One 6: e24570.
Acknowledgements
This work was supported by the Spanish Ministry of Science and Innovation through project Consolider-Ingenio Malaspina 2010 (CSD2008-00077). We thank our fellow scientists and the crew and ship scientists of the different cruise legs for smooth operation. We particularly thank E Borrull, C Díez, E Lara and D Vaqué for help in DNA sampling and the rest of the microbiologists for sharing data. We would also like to thank the Physical Oceanography Malaspina team B2 for the acquisition, processing and calibration of the CTD data and TS Catalá for the help on delineating water masses. Sequencing at the Joint Genome Intitute was supported by US Department of Energy Joint Genome Intitute 2011 Microbes Program grant CSP 602 grant to SGA. The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02–05CH11231. We also thank R Logares and the Barcelona Supercomputer Center for providing access to the MareNostrum Supercomputer (grants BCV- 2011-2-0003/3-0005, 2012-1-0006/2-0002 to R Logares). Additional funding was provided by Spanish Ministry of Science and Innovation grant CGL2011–26848/BOS MicroOcean PANGENOMICS and the Spanish Ministry of Economy and Competitivity grant MALASPINOMICS (CTM2011–15461-E). GS and FMCC held PhD JAE-Predoc (CSIC) and FPI (MICINN) fellowships respectively and SGA was supported by FP7-OCEAN-2011 project ‘Micro B3’.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Salazar, G., Cornejo-Castillo, F., Benítez-Barrios, V. et al. Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J 10, 596–608 (2016). https://doi.org/10.1038/ismej.2015.137
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.137
This article is cited by
-
Increased prokaryotic diversity in the Red Sea deep scattering layer
Environmental Microbiome (2023)
-
Abiotic selection of microbial genome size in the global ocean
Nature Communications (2023)
-
Top abundant deep ocean heterotrophic bacteria can be retrieved by cultivation
ISME Communications (2023)
-
A dataset of micro biodiversity in benthic sediment at a global scale
Scientific Data (2023)
-
Selection, drift and community interactions shape microbial biogeographic patterns in the Pacific Ocean
The ISME Journal (2022)