Abstract
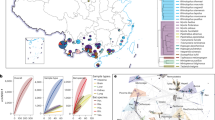
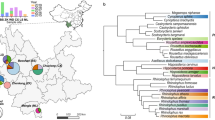
Studies have demonstrated that ~60%–80% of emerging infectious diseases (EIDs) in humans originated from wild life. Bats are natural reservoirs of a large variety of viruses, including many important zoonotic viruses that cause severe diseases in humans and domestic animals. However, the understanding of the viral population and the ecological diversity residing in bat populations is unclear, which complicates the determination of the origins of certain EIDs. Here, using bats as a typical wildlife reservoir model, virome analysis was conducted based on pharyngeal and anal swab samples of 4440 bat individuals of 40 major bat species throughout China. The purpose of this study was to survey the ecological and biological diversities of viruses residing in these bat species, to investigate the presence of potential bat-borne zoonotic viruses and to evaluate the impacts of these viruses on public health. The data obtained in this study revealed an overview of the viral community present in these bat samples. Many novel bat viruses were reported for the first time and some bat viruses closely related to known human or animal pathogens were identified. This genetic evidence provides new clues in the search for the origin or evolution pattern of certain viruses, such as coronaviruses and noroviruses. These data offer meaningful ecological information for predicting and tracing wildlife-originated EIDs.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A et al. (2013). A strategy to estimate unknown viral diversity in mammals. MBio 4: e00598–e00613.
Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM et al. (2014). Evidence for Camel-to-Human Transmission of MERS Coronavirus. N Engl J Med 370: 2499–2505.
Brown KE . (2010). The expanding range of parvoviruses which infect humans. Rev Med Virol 20: 231–244.
Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T . (2006) Bats: important reservoir hosts of emerging viruses Clin Microbiol Rev 19: 531–545.
Chae C . (2005). A review of porcine circovirus 2-associated syndromes and diseases. Vet J 169: 326–336.
Chen L, Liu B, Yang J, Jin Q . (2014). DBatVir: the database of bat-associated viruses. Database (Oxford) 2014: bau021.
Chinese SARS Molecular Epidemiology Consortium.. (2004). Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303: 1666–1669.
Chu DK, Poon LL, Gomaa MM, Shehata MM, Perera RA, Abu Zeid D et al. (2014). MERS Coronaviruses in Dromedary Camels, Egypt. Emerg Infect Dis 20: 1049–1053.
Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C et al. (2014a). Rooting the phylogenetic tree of MERS-Coronavirus by characterization of a conspecific virus from an African Bat. J Virol 88: 11297–11303.
Corman VM, Kallies R, Philipps H, Gopner G, Muller MA, Eckerle I et al. (2014b). Characterization of a novel betacoronavirus related to middle East respiratory syndrome coronavirus in European hedgehogs. J Virol 88: 717–724.
Cui J, Han N, Streicker D, Li G, Tang X, Shi Z et al. (2007). Evolutionary relationships between bat coronaviruses and their hosts. Emerg Infect Dis 13: 1526–1532.
Daszak P, Cunningham AA, Hyatt AD . (2000). Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287: 443–449.
Donaldson EF, Haskew AN, Gates JE, Huynh J, Moore CJ, Frieman MB . (2010). Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J Virol 84: 13004–13018.
Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T et al. (2012). Bats host major mammalian paramyxoviruses. Nat Commun 3: 796.
Fauquet CM, Fargette D . (2005). International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol J 2: 64.
Garcia-Perez R, Ibanez C, Godinez JM, Arechiga N, Garin I, Perez-Suarez G et al. (2014). Novel papillomaviruses in free-ranging Iberian bats: no virus-host co-evolution, no strict host specificity, and hints for recombination. Genome Biol Evol 6: 94–104.
Ge X, Li Y, Yang X, Zhang H, Zhou P, Zhang Y et al. (2012). Metagenomic analysis of viruses from the bat fecal samples reveals many novel viruses in insectivorous bats in China. J Virol 88: 4620–4630.
Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH et al. (2013). Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503: 535–538.
He B, Li Z, Yang F, Zheng J, Feng Y, Guo H et al. (2013). Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel Mammalian viruses. PLoS One 8: e61950.
He B, Zhang Y, Xu L, Yang W, Yang F, Feng Y et al. (2014). Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J Virol 88: 7070–7082.
Hemida MG, Al-Naeem A, Perera RA, Chin AW, Poon LL, Peiris M . (2015). Lack of Middle East respiratory syndrome coronavirus transmission from infected camels. Emerg Infect Dis 21: 699–701.
Holmes KV, Enjuanes L . (2003). Virology. The SARS coronavirus: a postgenomic era. Science 300: 1377–1378.
Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL et al. (2008). Global trends in emerging infectious diseases. Nature 451: 990–993.
Lau SK, Woo PC, Wong BH, Wong AY, Tsoi HW, Wang M et al. (2010). Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology 404: 106–116.
Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P et al. (2005). Fruit bats as reservoirs of Ebola virus. Nature 438: 575–576.
Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH et al. (2010a). Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84: 6955–6965.
Li L, McGraw S, Zhu K, Leutenegger CM, Marks SL, Kubiski S et al. (2013). Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect Dis 19: 534–541.
Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH et al. (2005). Bats are natural reservoirs of SARS-like coronaviruses. Science 310: 676–679.
Li Y, Ge X, Hon CC, Zhang H, Zhou P, Zhang Y et al. (2010b). Prevalence and genetic diversity of adeno-associated viruses in bats from China. J Gen Virol 91: 2601–2609.
Lloyd-Smith JO, George D, Pepin KM, Pitzer VE, Pulliam JR, Dobson AP et al. (2009). Epidemic dynamics at the human-animal interface. Science 326: 1362–1367.
Luis AD, Hayman DT, O'Shea TJ, Cryan PM, Gilbert AT, Pulliam JR et al. (2013). A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci 280: 20122753.
Marsh GA, Wang LF . (2012). Hendra and Nipah viruses: why are they so deadly? Curr Opin Virol 2: 242–247.
Mayo MA . (2002). A summary of taxonomic changes recently approved by ICTV. Arch Virol 147: 1655–1663.
Meyer B, Muller MA, Corman VM, Reusken CB, Ritz D, Godeke GJ et al. (2014). Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis 20: 552–559.
Namiki T, Hachiya T, Tanaka H, Sakakibara Y . (2012). MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res 40: e155.
Ng TF, Driscoll C, Carlos MP, Prioleau A, Schmieder R, Dwivedi B et al. (2013). Distinct lineage of vesiculovirus from big brown bats, United States. Emerg Infect Dis 19: 1978–1980.
O'Shea TJ, Cryan PM, Cunningham AA, Fooks AR, Hayman DT, Luis AD et al. (2014). Bat flight and zoonotic viruses. Emerg Infect Dis 20: 741–745.
Peng Y, Leung HC, Yiu SM, Chin FY . (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428.
Phan TG, Vo NP, Bonkoungou IJ, Kapoor A, Barro N, O'Ryan M et al. (2012). Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J Virol 86: 11024–11030.
Quan PL, Firth C, Street C, Henriquez JA, Petrosov A, Tashmukhamedova A et al. (2010). Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. MBio 1: e00208–e00210.
Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ et al. (2013). Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci USA 110: 8194–8199.
Smith I, Wang LF . (2013). Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol 3: 84–91.
Tracy S, Chapman NM, Drescher KM, Kono K, Tapprich W . (2006). Evolution of virulence in picornaviruses. Curr Top Microbiol Immunol 299: 193–209.
Wang X, Ren J, Gao Q, Hu Z, Sun Y, Li X et al. (2015). Hepatitis A virus and the origins of picornaviruses. Nature 517: 85–88.
Wolfe ND, Dunavan CP, Diamond J . (2007). Origins of major human infectious diseases. Nature 447: 279–283.
Woo PC, Lau SK, Huang Y, Yuen KY . (2009). Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood) 234: 1117–1127.
Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH et al. (2012). Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86: 3995–4008.
Wu Z, Ren X, Yang L, Hu Y, Yang J, He G et al. (2012). Virome analysis for identification of novel mammalian viruses in bat species from chinese provinces. J Virol 86: 10999–11012.
Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J et al. (2014). Novel henipa-like virus, mojiang paramyxovirus, in rats, China, 2012. Emerg Infect Dis 20.
Yahiro T, Wangchuk S, Tshering K, Bandhari P, Zangmo S, Dorji T et al. (2014). Novel human bufavirus genotype 3 in children with severe diarrhea, bhutan. Emerg Infect Dis 20: 1037–1039.
Yang J, Yang F, Ren L, Xiong Z, Wu Z, Dong J et al. (2011). Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J Clin Microbiol 49: 3463–3469.
Yang L, Wu Z, Ren X, Yang F, He G, Zhang J et al. (2013). Novel SARS-like betacoronaviruses in bats, China, 2011. Emerg Infect Dis 19.
Yang L, Wu Z, Ren X, Yang F, Zhang J, He G et al. (2014a). MERS-related betacoronavirus in Vespertilio superans bats, China. Emerg Infect Dis 20: 1260–1262.
Yang Y, Du L, Liu C, Wang L, Ma C, Tang J et al. (2014b). Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci USA 111: 12516–12521.
Acknowledgements
This study was supported by the Program for Changjiang Scholars and Innovative Research Team in University of China (IRT13007), the National S&T Major Project, ‘China Mega-Project for Infectious Disease’ (grant number 2011ZX10004-001 and 2014ZX10004001) from China and the PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (grant number 3332015095).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Wu, Z., Yang, L., Ren, X. et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J 10, 609–620 (2016). https://doi.org/10.1038/ismej.2015.138
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.138
This article is cited by
-
Selective deforestation and exposure of African wildlife to bat-borne viruses
Communications Biology (2024)
-
Identification of small circular DNA viruses in coyote fecal samples from Arizona (USA)
Archives of Virology (2024)
-
Metagenomic analysis of herbivorous mammalian viral communities in the Northwest Plateau
BMC Genomics (2023)
-
Comparative virome analysis of individual shedding routes of Miniopterus phillipsi bats inhabiting the Wavul Galge cave, Sri Lanka
Scientific Reports (2023)
-
A circovirus and cycloviruses identified in feces of bobcats (Lynx rufus) in California
Archives of Virology (2023)