Abstract
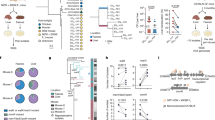
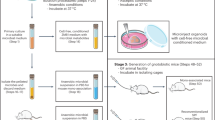
Endogenous intestinal microbiota have wide-ranging and largely uncharacterized effects on host physiology. Here, we used reverse-phase liquid chromatography-coupled tandem mass spectrometry to define the mouse intestinal proteome in the stomach, jejunum, ileum, cecum and proximal colon under three colonization states: germ-free (GF), monocolonized with Bacteroides thetaiotaomicron and conventionally raised (CR). Our analysis revealed distinct proteomic abundance profiles along the gastrointestinal (GI) tract. Unsupervised clustering showed that host protein abundance primarily depended on GI location rather than colonization state and specific proteins and functions that defined these locations were identified by random forest classifications. K-means clustering of protein abundance across locations revealed substantial differences in host protein production between CR mice relative to GF and monocolonized mice. Finally, comparison with fecal proteomic data sets suggested that the identities of stool proteins are not biased to any region of the GI tract, but are substantially impacted by the microbiota in the distal colon.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Cash H, Whitham C, Behrendt C, Hooper L . (2006). Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313: 1126–1130.
Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30: 918–920.
Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK . (2010). Host-bacterial symbiosis in health and disease. Adv Immunol 107: 243–274.
de Hoon MJ, Imoto S, Nolan J, Miyano S . (2004). Open source clustering software. Bioinformatics 20: 1453–1454.
Dimmer EC, Huntley RP, Alam-Faruque Y, Sawford T, O’Donovan C, Martin MJ et al. (2012). The UniProt-GO Annotation database in 2011. Nucleic Acids Res 40: D565–D570.
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M et al. (2005). Diversity of the human intestinal microbial flora. Science 308: 1635–1638.
Elias JE, Gygi SP . (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214.
3rd. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a aprotein database. J Am Soc Mass Spectrom 5: 976–989.
Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C et al. (2012). Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn’s disease. PLoS One 7: e49138.
Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR . (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785.
Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359.
Hooper LV, Falk PG, Gordon JI . (2000). Analyzing the molecular foundations of commensalism in the mouse intestine. Curr Opin Microbiol 3: 79–85.
Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA et al. (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189.
Ley RE, Turnbaugh PJ, Klein S, Gordon JI . (2006). Human gut microbes associated with obesity. Nature 444: 1022–1023.
Liaw A, Wiener M . (2002). Classification and regression by randomForest. R News 2: 18–22.
Lichtman JS, Marcobal A, Sonnenburg JL, Elias JE . (2013). Host-centric proteomics of stool: a novel strategy focused on intestinal responses to the gut microbiota. Mol Cell Proteomics 12: 3310–3318.
Lichtman JS, Sonnenburg JL, Elias JE . (2015). Monitoring host responses to the gut microbiota. ISME J 9: 1908–1915.
Lowe ME . (2002). The triglyceride lipases of the pancreas. J Lipid Res 43: 2007–2016.
Magurran AE . (2004) Measuring Biological Diversity 1st edn. Blackwell Sciences Ltd.: Malden, MA, USA.
Mahowald M a, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A et al. (2009). Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA 106: 5859–5864.
Martens EC, Chiang HC, Gordon JI . (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4: 447–457.
Maslowski KM, Mackay CR . (2011). Diet, gut microbiota and immune responses. Nat Immunol 12: 5–9.
Meeuwsen CG, De Visser H, Hazenberg MP, Wostmannn BS, Pleasantsa JR, Benner R et al. (1989). Serum immunoglobulin levels and naturally occurring antibodies against carbohydrate antigens in germ-free BALB/c mice fed chemically defined ultrafiltered diet. Eur J Immunol 19: 2335–2339.
Muth T, Kolmeder CA, Salojärvi J, Keskitalo S, Varjosalo M, Verdam FJ et al. (2015). Navigating through metaproteomics data: a logbook of database searching. Proteomics 15: 3439–3453.
Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F et al. (2007). Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut 56: 661–667.
Saldanha AJ . (2004). Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248.
Savage D . (1977). Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31: 107–133.
Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN et al. (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141: 1241–1252.
Suzuki K, Fagarasan S . (2008). How host-bacterial interactions lead to IgA synthesis in the gut. Trends Immunol 29: 523–531.
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI . (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031.
Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O et al. (2011). The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334: 255–258.
Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J et al. (2009). Shotgun metaproteomics of the human distal gut microbiota. ISME J 3: 179–189.
Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM et al. (2013). The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41: D1063–D1069.
Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T et al. (2011). Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11: 2019–2026.
Wen L, Ley R, Volchkov P, Stranges P . (2008). Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455: 1109–1113.
Whitman WB, Coleman DC, Wiebe WJ . (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583.
Zaia J . (2008). Mass spectrometry and the emerging field of glycomics. Chem Biol 15: 881–892.
Zhang H, Guo T, Li X, Datta A, Park JE, Yang J et al. (2010). Simultaneous characterization of glyco- and phosphoproteomes of mouse brain membrane proteome with electrostatic repulsion hydrophilic interaction chromatography. Mol Cell Proteomics 9: 635–647.
Acknowledgements
We would like to thank members of the Elias and Sonnenburg lab members, Carlos Gonzales and Katherine Ng for artistic support; and teaching assistants Alexandre Colavin, Miriam Gutschow and Sam Smits for helpful feedback. This work was supported by a Curriculum Development Award from The MathWorks, Inc. (to KCH and JLS), a grant from the National Institutes of Health (R01-DK085025) (to JLS), the Stanford Systems Biology Center funded by National Institutes of Health grant P50 GM107615 (to KCH) and grant DGE-114747 from the National Science Foundation (to JSL).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Lichtman, J., Alsentzer, E., Jaffe, M. et al. The effect of microbial colonization on the host proteome varies by gastrointestinal location. ISME J 10, 1170–1181 (2016). https://doi.org/10.1038/ismej.2015.187
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.187
This article is cited by
-
DeepRTAlign: toward accurate retention time alignment for large cohort mass spectrometry data analysis
Nature Communications (2023)
-
Maternal milk and fecal microbes guide the spatiotemporal development of mucosa-associated microbiota and barrier function in the porcine neonatal gut
BMC Biology (2019)
-
The Interplay Between the Microbiome and Cardiovascular Risk
Current Genetic Medicine Reports (2018)
-
Metaproteomic analysis of human gut microbiota: where are we heading?
Journal of Biomedical Science (2017)
-
The effect of microbial challenge on the intestinal proteome of broiler chickens
Proteome Science (2016)