Abstract
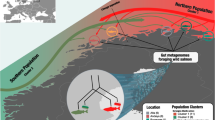
Although understood in many vertebrate systems, the natural diversity of host-associated microbiota has been little studied in teleosts. For migratory fishes, successful exploitation of multiple habitats may affect and be affected by the composition of the intestinal microbiome. We collected 96 Salmo salar from across the Atlantic encompassing both freshwater and marine phases. Dramatic differences between environmental and gut bacterial communities were observed. Furthermore, community composition was not significantly impacted by geography. Instead life-cycle stage strongly defined both the diversity and identity of microbial assemblages in the gut, with evidence for community destabilisation in migratory phases. Mycoplasmataceae phylotypes were abundantly recovered in all life-cycle stages. Patterns of Mycoplasmataceae phylotype recruitment to the intestinal microbial community among sites and life-cycle stages support a dual role for deterministic and stochastic processes in defining the composition of the S. salar gut microbiome.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bruno D, Noguera P, Poppe T . (2013) A Colour Atlas of Salmonid Diseases. Springer: Heidelberg, Germany.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Freitas S, Hatosy S, Fuhrman JA, Huse SM, Mark Welch DB, Sogin ML et al. (2012). Global distribution and diversity of marine Verrucomicrobia. ISME J 6: 1499–1505.
Frey J, Herrmann R . (2002) Mycoplasmas of animals. In: Razin S, Herrmann R (eds). Molecular Biology and Pathogenicity of Mycoplasmas. Kluwer: New York, USA.
Green TJ, Smullen R, Barnes AC . (2013). Dietary soybean protein concentrate-induced intestinal disorder in marine farmed Atlantic salmon, Salmo salar is associated with alterations in gut microbiota. Vet Microbiol 166: 286–292.
Holben WE, Williams P, Gilbert M, Saarinen M, Sarkilahti LK, Apajalahti JH . (2002). Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Microb Ecol 44: 175–185.
Jacobsen J, Hansen L . (1999) Feeding habits of Atlantic salmon at different life stages at sea. In: Mills D (ed). The Ocean Life of Atlantic Salmon: Environmental and Biological Factors Influencing Survival. Wiley-Blackwell: London, UK.
Llewellyn M, Boutin S, Hoseinifar SH, Derome N . (2014). Teleost microbiomes: progress towards their characterisation, manipulation and applications in aquaculture and fisheries. Front Microbiol 5: 207.
McCormick SD, Sheehan TF, Björnsson BT, Lipsky C, Kocik JF, Regish AM et al. (2013). Physiological and endocrine changes in Atlantic salmon smolts during hatchery rearing, downstream migration, and ocean entry. Can J Fisheries Aquat Sci 70: 105–118.
Nechitaylo TY, Timmis KN, Golyshin PN . (2009). ‘Candidatus Lumbricincola’, a novel lineage of uncultured Mollicutes from earthworms of family Lumbricidae. Environ Microbiol 11: 1016–1026.
Oksanen J, Blanchet F, Kindt K, Legendre P, Minchin P, O'Hara R et al. (2015), vegan: Community Ecology Package. R package version 2.2-1. http://CRAN.R-project.org/package=vegan.
Orlov AV, Gerasimov YV, Lapshin OM . (2006). The feeding behaviour of cultured and wild Atlantic salmon, Salmo salar L., in the Louvenga River, Kola Peninsula, Russia. ICES J Mar Sci 63: 1297–1303.
Rawls JF, Mahowald MA, Ley RE, Gordon JI . (2006). Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127: 423–433.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Schmidt VT, Smith KF, Melvin DW, Amaral-Zettler LA . (2015). Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol Ecol 24: 2537–2550.
Star B, Haverkamp THA, Jentoft S, Jakobsen KS . (2013). Next generation sequencing shows high variation of the intestinal microbial species composition in Atlantic cod caught at a single location. BMC Microbiol 13: 248–248.
Stefansson S, Bjornsson B, Ebbensson L, McCormick S . (2008) Smoltification. In: Kappor BJ, Finn RN (eds). Fish Larval Physiology. Wiley-Blackwell: London, UK.
Sullam KE, Essinger SD, Lozupone CA, O'Connor MP, Rosen GL, Knight R et al. (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol 21: 3363–3378.
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M et al. (2012). Human gut microbiome viewed across age and geography. Nature 486: 222–227.
Zarkasi KZ, Abell GCJ, Taylor RS, Neuman C, Hatje E, Tamplin ML et al. (2014). Pyrosequencing-based characterization of gastrointestinal bacteria of Atlantic salmon (Salmo salar L.) within a commercial mariculture system. J Appl Microbiol 117: 18–27.
Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y et al. (2009). Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci 106: 2365–2370.
Acknowledgements
We thank the assistance of Julien April and Denise Deschamps, Ministère des Ressources Naturelles et de la Faune, Quebec Canada; Tim Sheehan, NOAA Fisheries Service; Woods Hole, USA; Ger Rogan, Russell Poole, Katie Thomas, Niall O’Maoileidigh, Deirdre Cotter and Elvira de Eyto, Marine Institute, Ireland; Paddy Gargan and Michael Hughes, Inland Fisheries Ireland; and Brian Clarke and Tom Reed, University College Cork in sample collection. MSL was funded by a Marie Skłodowska-Curie International Outgoing Fellowship, grant number 302503. JL was funded by a NSERC-CREATE Fellowship. FT was funded by a NSERC discovery grant awarded to ND. PMcG was supported by the Beaufort Marine Fish Award in Fish Population Genetics funded by the Irish Government under the Sea Change Programme.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Llewellyn, M., McGinnity, P., Dionne, M. et al. The biogeography of the atlantic salmon (Salmo salar) gut microbiome. ISME J 10, 1280–1284 (2016). https://doi.org/10.1038/ismej.2015.189
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.189
This article is cited by
-
Impact of the diet in the gut microbiota after an inter-species microbial transplantation in fish
Scientific Reports (2024)
-
Host species and habitat shape fish-associated bacterial communities: phylosymbiosis between fish and their microbiome
Microbiome (2023)
-
Effect of yeast species and processing on intestinal microbiota of Atlantic salmon (Salmo salar) fed soybean meal-based diets in seawater
Animal Microbiome (2023)
-
Host-microbiota-parasite interactions in two wild sparid fish species, Diplodus annularis and Oblada melanura (Teleostei, Sparidae) over a year: a pilot study
BMC Microbiology (2023)
-
Bacterial networks in Atlantic salmon with Piscirickettsiosis
Scientific Reports (2023)