Abstract
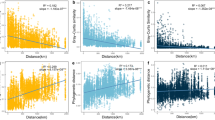
Bacteria play key roles in the ecology of both aquatic and terrestrial ecosystems; however, little is known about their diversity and biogeography, especially in the rare microbial biosphere of inland freshwater ecosystems. Here we investigated aspects of the community ecology and geographical distribution of abundant and rare bacterioplankton using high-throughput sequencing and examined the relative influence of local environmental variables and regional (spatial) factors on their geographical distribution patterns in 42 lakes and reservoirs across China. Our results showed that the geographical patterns of abundant and rare bacterial subcommunities were generally similar, and both of them showed a significant distance–decay relationship. This suggests that the rare bacterial biosphere is not a random assembly, as some authors have assumed, and that its distribution is most likely subject to the same ecological processes that control abundant taxa. However, we identified some differences between the abundant and rare groups as both groups of bacteria showed a significant positive relationship between sites occupancy and abundance, but the abundant bacteria exhibited a weaker distance–decay relationship than the rare bacteria. Our results implied that rare subcommunities were mostly governed by local environmental variables, whereas the abundant subcommunities were mainly affected by regional factors. In addition, both local and regional variables that were significantly related to the spatial variation of abundant bacterial community composition were different to those of rare ones, suggesting that abundant and rare bacteria may have discrepant ecological niches and may play different roles in natural ecosystems.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Baas-Becking LGM. (1934) Geobiologie of Inleiding Tot de Milieukunde. WP Van Stockum & Zoon: The Hague: The Netherlands.
Barberán A, Casamayor EO . (2010). Global phylogenetic community structure and β-diversity patterns in surface bacterioplankton metacommunities. Aquat Microb Ecol 59: 1–10.
Berdjeb L, Ghiglione JF, Jacquet S . (2011). Bottom-up versus top-down control of hypo- and epilimnion free-living bacterial community structures in two neighboring freshwater lakes. Appl Environ Microbiol 77: 3591–3599.
Blanchet FG, Legendre P, Borcard D . (2008). Forward selection of explanatory variables. Ecology 89: 2623–2632.
Borcard D, Legendre P . (2002). All-scale spatial analysis of ecological data by means of principal coordinates of neighbor matrices. Ecol Model 153: 51–68.
Brown MV, Ostrowski M, Grzymski J, Lauro FM . (2014). A trait based perspective on the biogeography of common and abundant marine bacterioplankton clades. Mar Genom 15: 17–28.
Caughley G . (1994). Directions in conservation biology. J Animal Ecol 63: 215–244.
Cavender-Bares J, KozakK H, Fine PVA, Kembel SW . (2009). The merging of community ecology and phylogenetic biology. Ecol Lett 12: 693–715.
Claesson MJ, O’Sullivan O, Wang Q, Nikkila J, Marchesi JR, Smidt H et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 8: e6669.
Clarke KR, Gorley RN . (2001) PRIMER v5: User Manual/Tutorial. PRIMER-E: Plymouth: UK.
Cottenie K . (2005). Integrating environmental and spatial processes in ecological community dynamics. Ecol Lett 8: 1175–1182.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R . (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200.
Fenchel T . (2003). Biogeography for bacteria. Science 301: 925–927.
Fenchel T, Esteban GF, Finlay BJ . (1997). Local versus global diversity of microorganisms: cryptic diversity of ciliate protozoa. Oikos 80: 220–225.
Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631.
Figuerola J, Green AJ, Michot TC . (2005). Invertebrate eggs can fly: evidence of waterfowl-mediated gene flow in aquatic invertebrates. Am Nat 165: 274–280.
Finlay BJ . (1998). The global diversity of protozoa and other small species. Int J Paristol 28: 29–48.
Finlay BJ . (2002). Global dispersal of free-living microbial eukaryote species. Science 296: 1061–1063.
Galand PE, Casamayor EO, Kirchman DL, Lovejoy C . (2009). Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA 106: 22427–22432.
Gaston KJ . (1994) Rarity. Chapman & Hall: London, UK.
Gaston KJ, Blackburn TM, Greenwood JJD, Gregory RD, Quinn RM, Lawton JH . (2000). Abundance-occupancy relationships. J Appl Ecol 37: 39–59.
Gobet A, Quince C, Ramette A . (2010). Multivariate cut off level analysis (MultiCoLA) of large community datasets. Nucleic Acids Res 38: e155.
Graham CH, Fine PVA . (2008). Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecol Lett 11: 1265–1277.
Green J, Bohannan BJM . (2006). Spatial scaling of microbial biodiversity. Trends Ecol Evol 21: 501–507.
Greenberg AE, Clesceri LS, Eaton AD . (1992) Standard Methods for the Examination of Water and Wastewater. American Public Health Association: Washington (DC), USA.
Grime JP . (1998). Benefits of plant diversity to ecosystems: intermediate, filter and founder effects. J Ecol 86: 902–910.
Hambright KD, Beyer JE, Easton JD, Zamor RM, Easton AC, Hallidayschult TC . (2015). The niche of an invasive marine microbe in a subtropical freshwater impoundment. ISME J 9: 256–264.
Hamilton WD, Lenton TM . (1998). Spora and Gaia: how microbes fly with their clouds. Etho Ecol Evol 10: 1–16.
Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH . (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10: 497–506.
Holt AR, Warren PH, Gaston KJ . (2004). The importance of habitat heterogeneity, biotic interactions and dispersal in abundance–occupancy relationships. J Anim Ecol 73: 841–851.
Hutchinson GE . (1964). The lacustrine microcosm reconsidered. Am Sci 52: 334–341.
Jones SE, Cadkin TA, Newton RJ, McMahon KD . (2012). Spatial and temporal scales of aquatic bacterial beta diversity. Front Microbiol 3: 1–10.
Ju LH, Yang J, Liu LM, Wilkinson DM . (2014). Diversity and distribution of freshwater testate amoebae (Protozoa) along latitudinal and trophic gradients in China. Microb Ecol 68: 657–670.
Judd KE, Crump BC, Kling GW . (2006). Variation in dissolved organic matter controls bacterial production and community composition. Ecology 87: 2068–2079.
Kent M . (2012) Vegetation Description and Data Analysis, 2nd edn. Wiley-Blackwell: Chichester, UK.
Kim T, Jeong J, Wells GF, Park H . (2013). General and rare bacterial taxa demonstrating different temporal dynamic patterns in an activated sludge bioreactor. Appl Microbiol Biotechnol 97: 1755–1765.
Lear G, Bellamy J, Case BS, Lee JE, Buckley HL . (2014). Fine-scale spatial patterns in bacterial community composition and function within freshwater ponds. ISME J 8: 1715–1726.
Legendre P, Bordard D, Peres-Neto P . (2008). Analyzing or explaining betadiversity? Comment. Ecology 89: 3238–3244.
Legendre P, Legendre L . (2012) Numerical Ecology, 3rd edn. Elsevier: Amsterdam: The Netherlands.
Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF et al. (2004). The metacommunity concept: a frame work for multi-scale community ecology. Ecol Lett 7: 601–613.
Lepš J, Šmilauer P . (2003) Multivariate Analysis of Ecological Data Using CANOCO. Cambridge University Press: Cambridge, UK.
Lindström ES, Langenheder S . (2012). Local and regional factors influencing bacterial community assembly. Environ Microbiol Rep 4: 1–9.
Liu LM, Yang J, Yu XQ, Chen GJ, Yu Z . (2013). Patterns in the composition of microbial communities from a subtropical river: effects of environmental, spatial and temporal factors. PLoS One 8: e81232.
Logares R, Lindström ES, Langenheder S, Logue JB, Paterson H, Laybourn-Parry J et al. (2013). Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J 7: 937–948.
Logares R, Audic S, Bass D, Bittner L, Boutte C, Christen R et al. (2014). Patterns of rare and abundant marine microbial eukaryotes. Curr Biol 24: 813–821.
Logue JB, Lindström ES . (2008). Biogeography of bacterioplankton in inland waters. Freshw Rev 1: 99–114.
Logue JB . (2010) Factors influencing the biogeography of bacteria in freshwaters—a metacommunity approach. PhD thesis, Uppsala University. Uppsala: Sweden.
Magurran AE, Henderson PA . (2003). Explaining the excess of rare species in natural species abundance distributions. Nature 422: 714–716.
Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102–112.
Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK et al. (2011). Global patterns in the biogeography of bacterial taxa. Environ Microbiol 13: 135–144.
Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S . (2011). A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75: 14–49.
Pedrós-Alió C . (2006). Marine microbial diversity: can it be determined? Trends Microbiol 14: 257–263.
Pedrós-Alió C . (2012). The rare bacterial biosphere. Annu Rev Mar Sci 4: 449–466.
Pernthaler J . (2005). Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3: 537–546.
Pester M, Bittner N, Deevong P, Wagner M, Loy A . (2010). A ‘rare biosphere’ microorganism contributes to sulfate reduction in a peatland. ISME J 4: 1591–1602.
Preston FW . (1948). The commonness and rarity of species. Ecology 29: 254–283.
Ramette A, Tiedje JM . (2007). Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol 53: 197–207.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Teira E, Lekunberri I, Gasol JM, Nieto-Cid M, Álvarez-Salgado XA, Figueiras PG . (2007). Dynamics of the hydrocarbon-degrading Cycloclasticus bacteria during mesocosm-simulated oil spills. Environ Microbiol 9: 2551–2562.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Wang J, Shen J, Wu Y, Tu C, Soininen J, Stegen JC et al. (2013). Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. ISME J 7: 1310–1321.
Wilkinson DM, Koumoutsaris S, Mitchell EAD, Bey I . (2012). Modelling the effects of size on the aerial dispersal of microorganisms. J Biogeogr 39: 89–97.
Acknowledgements
We thank Zhengwen Liu, Kuanyi Li, Feizhou Chen and Bin Xue at Nanjing Institute of Geography and Limnology, Chinese Academy of Sciences, for field sampling assistance and providing part of physicochemical data. This work was funded by the National Basic Research Program of China (2012CB956103), the National Natural Science Foundation of China (31370471), the Natural Science Foundation for Distinguished Young Scholars of Fujian Province (2012J06009) and the Knowledge Innovation Program of the Chinese Academy of Sciences (IUEQN201305). We thank our referees for insightful comments that helped in improving the clarity of this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, L., Yang, J., Yu, Z. et al. The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J 9, 2068–2077 (2015). https://doi.org/10.1038/ismej.2015.29
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.29
This article is cited by
-
Stochastic community assembly of abundant taxa maintains the relationship of soil biodiversity-multifunctionality under mercury stress
Soil Ecology Letters (2024)
-
Spatial distribution pattern across multiple microbial groups along an environmental stress gradient in tobacco soil
Annals of Microbiology (2023)
-
Core and conditionally rare taxa as indicators of agricultural drainage ditch and stream health and function
BMC Microbiology (2023)
-
Vegetation as a key driver of the distribution of microbial generalists that in turn shapes the overall microbial community structure in the low Arctic tundra
Environmental Microbiome (2023)
-
Assembly processes and functional diversity of marine protists and their rare biosphere
Environmental Microbiome (2023)