Abstract
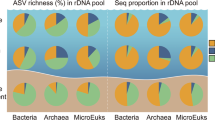
Hypersaline meromictic lakes are extreme environments in which water stratification is associated with powerful physicochemical gradients and high salt concentrations. Furthermore, their physical stability coupled with vertical water column partitioning makes them important research model systems in microbial niche differentiation and biogeochemical cycling. Here, we compare the prokaryotic assemblages from Ursu and Fara Fund hypersaline meromictic lakes (Transylvanian Basin, Romania) in relation to their limnological factors and infer their role in elemental cycling by matching taxa to known taxon-specific biogeochemical functions. To assess the composition and structure of prokaryotic communities and the environmental factors that structure them, deep-coverage small subunit (SSU) ribosomal RNA (rDNA) amplicon sequencing, community domain-specific quantitative PCR and physicochemical analyses were performed on samples collected along depth profiles. The analyses showed that the lakes harbored multiple and diverse prokaryotic communities whose distribution mirrored the water stratification patterns. Ursu Lake was found to be dominated by Bacteria and to have a greater prokaryotic diversity than Fara Fund Lake that harbored an increased cell density and was populated mostly by Archaea within oxic strata. In spite of their contrasting diversity, the microbial populations indigenous to each lake pointed to similar physiological functions within carbon degradation and sulfate reduction. Furthermore, the taxonomy results coupled with methane detection and its stable C isotope composition indicated the presence of a yet-undescribed methanogenic group in the lakes’ hypersaline monimolimnion. In addition, ultrasmall uncultivated archaeal lineages were detected in the chemocline of Fara Fund Lake, where the recently proposed Nanohaloarchaeota phylum was found to thrive.
Similar content being viewed by others

Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
Accessions
Sequence Read Archive
References
Alexe M . (2010) Studiul Lacurilor Sărate Din Depresiunea Transilvaniei ed Presa Universitară Clujeană: Cluj-Napoca, Romania, (in Romanian).
Andrei A-Ş, Banciu HL, Oren A . (2012). Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol Lett 330: 1–9.
Auguet JC, Triado-Margarit X, Nomokonova N, Camarero L, Casamayor EO . (2012). Vertical segregation and phylogenetic characterization of ammonia-oxidizing Archaea in a deep oligotrophic lake. ISME J 6: 1786–1797.
Baati H, Guermazi S, Gharsallah N, Sghir A, Ammar E . (2010). Novel prokaryotic diversity in sediments of Tunisian multipond solar saltern. Res Microbiol 161: 573–582.
Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD et al. (2010). Enigmatic, ultrasmall, uncultivated archaea. Proc Natl Acad Sci USA 107: 8806–8811.
Barberán A, Casamayor EO . (2011). Euxinic freshwater hypolimnia promote bacterial endemicity in continental areas. Microb Ecol 61: 465–472.
Baricz A, Coman C, Andrei A-Ş, Muntean V, Keresztes ZG, Păusan M et al. (2014). Spatial and temporal distribution of archaeal diversity in meromictic, hypersaline Ocnei Lake (Transylvanian Basin, Romania). Extremophiles 18: 399–413.
Benlloch S, López-López A, Casamayor EO, Øvreås L, Goddard V, Daae FL et al. (2002). Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol 4: 349–360.
Biderre-Petit C, Jézéquel D, Dugat-Bony E, Lopes F, Kuever J, Borrel G et al. (2011). Identification of microbial communities involved in the methane cycle of a freshwater meromictic lake. FEMS Microbiol Ecol 77: 533–545.
Boehrer B, Schultze M . (2009). Density stratification and stability. In: Likens GE (ed) Encyclopedia of Inland Waters. Elsevier: Oxford, pp 583–593.
Bonin AS, Boone DR . (2006). The order Methanobacteriales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The Prokaryotes, Volume 3: Archaea. Bacteria: Firmicutes, Actinomycetes. Springer-Verlag New York, NY, USA, pp 231–243.
Bowman JP, McCammon SA, Rea SM, McMeekin TA . (2000). The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol Lett 183: 81–88.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010a). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R . (2010b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267.
Cappellen PV, Viollier E, Roychoudhury A . (1998). Biogeochemical cycles of manganese and iron at the oxic-anoxic transition of a stratified marine basin (Orca Basin, Gulf of Mexico). Environ Sci Technol 32: 2931–2939.
Clesceri LS, Greenberg AE, Eaton AD . (1999) Standard Methods for the Examination of Water and Wastewater, 20th edn. APHA, AWWA, WEF: Washington, USA.
Comeau AM, Harding T, Galand PE, Vincent WF, Lovejoy C . (2012). Vertical distribution of microbial communities in a perennially stratified Arctic lake with saline, anoxic bottom waters. Sci Rep 2: 604.
Cristea A, Andrei A-Ş, Baricz A, Muntean V, Banciu HL . (2014). Rapid assessment of carbon substrate utilization in the epilimnion of meromictic Ursu Lake (Sovata, Romania) by the BIOLOG EcoplateTM approach. Studia UBB Biologia 59: 41–53.
Demergasso C, Escudero L, Casamayor EO, Chong G, Balagué V, Pedrós-Alió C . (2008). Novelty and spatio-temporal heterogeneity in the bacterial diversity of hypersaline Lake Tebenquiche (Salar de Atacama). Extremophiles 12: 491–504.
Edberg F, Andersson AF, Holmström SJ . (2012). Bacterial community composition in the water column of a lake formed by a former uranium open pit mine. Microb Ecol 64: 870–880.
Edgar RC . (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998.
Einen J, Thorseth IH, Ovreås L . (2008). Enumeration of Archaea and Bacteria in seafloor basalt using real-time quantitative PCR and fluorescence microscopy. FEMS Microbiol Lett 282: 182–187.
Etiope G, Feyzullayev A, Baciu CL . (2009). Terrestrial methane seeps and mud volcanoes: a global perspective of gas origin. Mar Petrol Geol 26: 333–344.
Ferrer M, Werner J, Chernikova TN, Bargiela R, Fernández L, La Cono V et al. (2012). Unveiling microbial life in the new deep-sea hypersaline Lake Thetis. Part II: a metagenomic study. Environ Microbiol 14: 268–281.
Frontier S . (1985). Diversity and structure in aquatic ecosystems. Oceanogr Mar Biol 23: 253–312.
Ghai R, Pašić L, Fernández AB, Martin-Cuadrado A-B, Mizuno CM, McMahon KD et al. (2011). New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1: 135.
Habicht KS, Miller M, Cox RP, Frigaard N-U, Tonolla M, Peduzzi S et al. (2011). Comparative proteomics and activity of a green sulfur bacterium through the water column of Lake Cadagno, Switzerland. Environ Microbiol 13: 203–215.
Hamilton TL, Bovee RJ, Thiel V, Sattin SR, Mohr W, Schaperdoth I et al. (2014). Coupled reductive and oxidative sulfur cycling in the phototrophic plate of a meromictic lake. Geobiology 12: 451–468.
Hammer UT . (1986) Saline Lake Ecosystems of the World vol. 59. Dr W Junk Publishers: Dordrecht, The Netherlands, p 15.
Har N, Rusz O, Codrea V, Barbu O . (2010). New data on the mineralogy of the salt deposit from Sovata (Mureş County-Romania). Carpath J Earth Env 5: 127–135.
Hollister EB, Engledow AS, Hammett AJM, Provin TL, Wilkinson HH, Gentry TJ . (2010). Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J 4: 829–838.
Humayoun SB, Bano N, Hollibaugh JT . (2003). Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl Environ Microbiol 69: 1030–1042.
Hurtgen MT, Lyons TW, Ingall ED, Cruse AM . (1999). Anomalous enrichments of iron monosulfide in euxinic marine sediments and the role of H2S in iron sulfide transformations: examples from Effingham Inlet, Orca Basin, and the Black Sea. Am J Sci 299: 556–588.
Huson DH, Scornavacca C . (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61: 1061–1067.
Ionescu A . (2015). Geogenic methane in petroliferous and geothermal areas in Romania: origin and emission to the atmosphere. PhD thesis, Faculty of Environmental Science and Engineering. Babes-Bolyai University: Cluj-Napoca.
Iversen N, Oremland RS, Klug MJ . (1987). Big Soda Lake (Nevada).3. Pelagic methanogenesis and anaerobic methane oxidation. Limnol Oceanogr 32: 804–814.
Kelley CA, Poole JA, Tazaz AM, Chanton JP, Bebout BM . (2012). Substrate limitation for methanogenesis in hypersaline environments. Astrobiology 12: 89–97.
Keresztes ZG, Felföldi T, Somogyi B, Székely G, Dragoş N, Márialiget K et al. (2012). First record of picophytoplankton diversity in Central European hypersaline lakes. Extremophiles 16: 759–769.
La Cono V, La Spada G, Arcadi E, Placenti F, Smedile F, Ruggeri G et al. (2013). Partaking of Archaea to biogeochemical cycling in oxygen-deficient zones of meromictic saline Lake Faro (Messina, Italy). Environ Microbiol 15: 1717–1733.
Lane DJ . (1991). 16S/23S rRNA sequencing. In Stackebrandt E, Goodfellow M (eds) Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons Ltd: United Kingdom, pp 115–175.
Lauro FM, DeMaere MZ, Yau S, Brown MV, Ng C, Wilkins D et al. (2011). An integrative study of a meromictic lake ecosystem in Antarctica. ISME J 5: 879–895.
Lentini V, Gugliandolo C, Maugeri TL . (2012). Vertical distribution of Archaea and Bacteria in a meromictic lake as determined by fluorescent in situ hybridization. Curr Microbiol 64: 66–74.
Lepère C, Masquelier S, Mangot J-F, Debroas D, Domaizon I . (2010). Vertical structure of small eukaryotes in three lakes that differ by their trophic status: a quantitative approach. ISME J 4: 1509–1519.
Lopes F, Viollier E, Thiam A, Michard G, Abril G, Groleau A et al. (2011). Biogeochemical modeling of anaerobic vs. aerobic methane oxidation in a meromictic crater lake (Lake Pavin, France). Appl Geochem 26: 1919–1932.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL . (2013). Practical innovations for high-throughput amplicon sequencing. Nat Methods 10: 999–1002.
Marteinsson VT, Rúnarsson Á, Stefánsson A, Thorsteinsson T, Jóhannesson T et al. (2013). Microbial communities in the subglacial waters of the Vatnajökull ice cap, Iceland. ISME J 7: 427–437.
Martin M . (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12.
Máthé I, Borsodi AK, Tóth EM, Felföldi T, Jurecska L, Krett G et al. (2014). Vertical physico-chemical gradients with distinct microbial communities in the hypersaline and heliothermal Lake Ursu (Sovata, Romania). Extremophiles 18: 501–514.
McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D et al. (2012a). The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience 1: 7.
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012b). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618.
Muyzer G, Waal EC, Uitterlinden AG . (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700.
Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ et al. (2012). De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6: 81–93.
Oren A . (2011). Thermodynamic limits to microbial life at high salt concentrations. Environ Microbiol 13: 1908–1923.
Oren A . (2012). Approaches toward the study of halophilic microorganisms in their natural environments: who are they and what are they doing? In: Vreeland RH (ed) Advances in Understanding the Biology of Halophilic Microorganisms. Springer: New York, pp 1–33.
Pasche N, Dinkel C, Müller B, Schmid M, Wüest A, Wehrli B . (2009). Physical and biogeochemical limits to internal nutrient loading of meromictic Lake Kivu. Limnol Oceanogr 54: 1863–1873.
Pouliot J, Galand PE, Lovejoy C, Vincent WF . (2009). Vertical structure of archaeal communities and the distribution of ammonia monooxygenase A gene variants in two meromictic High Arctic lakes. Environ Microbiol 11: 687–699.
Price MN, Dehal PS, Arkin AP . (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490.
Pruesse E, Peplies J, Glöckner FO . (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: (Database issue) D590–D596.
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437.
Roden EE, Lovley DR . (1993). Dissimilatory Fe(III) reduction by the marine microorganism Desulfuromonas acetoxidans. Appl Environ Microbiol 59: 734–742.
Schubert CJ, Lucas FS, Durisch-Kaiser E, Stierli R, Diem T, Scheidegger O et al. (2010). Oxidation and emission of methane in a monomictic lake (Rotsee, Switzerland). Aquat Sci 72: 455–466.
Somogyi B, Vörös L, Pálffy K, Székely G, Bartha C, Keresztes ZG . (2014). Picophytoplankton predominance in hypersaline lakes (Transylvanian Basin, Romania). Extremophiles 18: 1075–1084.
Sorokin DY, Foti M, Tindall BJ, Muyzer G . (2007). Desulfurispirillum alkaliphilum gen. nov. sp. nov., a novel obligately anaerobic sulfur-and dissimilatory nitrate-reducing bacterium from a full-scale sulfide-removing bioreactor. Extremophiles 11: 363–370.
Sorokin DY, Tourova TP, Galinski EA, Muyzer G, Kuenen JG . (2008). Thiohalorhabdus denitrificans gen. nov., sp. nov., an extremely halophilic, sulfur-oxidizing, deep-lineage gammaproteobacterium from hypersaline habitats. Int J Syst Evol Microbiol 58: 2890–2897.
Spulber L, Etiope G, Baciu C, Malos C, Vlad SN . (2010). Methane emission from natural gas seeps and mud volcanoes in Transylvania (Romania). Geofluids 10: 463–475.
Tang XM, Gao G, Qin B, Zhu L, Chao J, Yang G . (2009). Characterization of bacterial communities associated with organic aggregates in a large, shallow, eutrophic freshwater lake (Lake Taihu, China). Microb Ecol 58: 307–322.
Trüper HG, Schlegel HG . (1964). Sulfur metabolism in Thiorhodaceae. 1. Quantitative measurements on growing cells of Chromatium okenii. Antonie van Leeuwenhoek 30: 225–238.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG et al. (2011). Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J 6: 94–103.
Wetzel RG, Likens GE . (1991) Limnological Analyses. Springer-Verlag: New York, p 39.
Wetzel RG, Likens GE . (1995) Limnological Analysis, 2nd edn. Springer-Verlag: New York, NY, USA.
Whiticar MJ, Faber E, Schoell M . (1986). Biogenic methane formation in marine and freshwater environments: CO2 reduction vs acetate fermentation – isotope evidence. Geochim Cosmochim Acta 50: 693–709.
Yakimov MM, La Cono V, Slepak VZ, La Spada G, Arcadi E, Messina E et al. (2013). Microbial life in the Lake Medee, the largest deep-sea salt-saturated formation. Sci Rep 3: 3554.
Yau S, Lauro FM, Williams TJ, Demaere MZ, Brown MV, Rich J et al. (2013). Metagenomic insights into strategies of carbon conservation and unusual sulfur biogeochemistry in a hypersaline Antarctic lake. ISME J 7: 1944–1961.
Zhaxybayeva O, Stepanauskas R, Mohan NR, Papke RT . (2013). Cell sorting analysis of geographically separated hypersaline environments. Extremophiles 17: 265–275.
Acknowledgements
This work was supported by grants of the Romanian National Authority for Scientific Research, CNCS–UEFIS-CDI, project numbers PN-II-ID-PCE-2011-3-0546 and PN-II-ID-PCE-2011-3-0765. A-ŞA was supported by a POSDRU/159/1.5/S/132400 research scholarship; CC was supported by Grants PN 09-360201 and POSDRU/159/1.5/S/133391; AI was supported by a POSDRU/159/1.5/S/133391 doctoral scholarship; MSR and MP were supported by Oak Ridge National Laboratory (ORNL). ORNL is managed by UT-Battelle, LLC, for the US Department of Energy. We thank Zamin Yang and Dawn Klingeman for the help provided with Illumina amplicon preparation and sequencing. We are grateful to Tudor Tămaş for the mineralogical analysis, Zsolt G Keresztes for supporting unpublished data and to Dimitry Y Sorokin for his critical review of the manuscript. We are grateful to Daniela Buta (Ocna Sibiului) and Nagy Fülop János (Sovata) for the permission to enter the study areas.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Andrei, AŞ., Robeson, M., Baricz, A. et al. Contrasting taxonomic stratification of microbial communities in two hypersaline meromictic lakes. ISME J 9, 2642–2656 (2015). https://doi.org/10.1038/ismej.2015.60
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.60
This article is cited by
-
Hydrography of Crater Lake of Isabel Island, Nayarit, México
Limnology (2022)
-
A comparison of genetically and morphometrically identified macroinvertebrate community index scores with implications for aquatic life use attainment
Environmental Monitoring and Assessment (2022)
-
Anion-type modulates the effect of salt stress on saline lake bacteria
Extremophiles (2022)
-
Comprehensive mineralogical and physicochemical characterization of recent sapropels from Romanian saline lakes for potential use in pelotherapy
Scientific Reports (2021)
-
Distinctive distributions of halophilic Archaea across hypersaline environments within the Qaidam Basin of China
Archives of Microbiology (2021)