Abstract
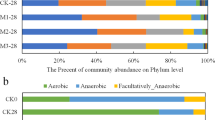
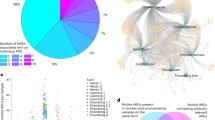
The overuse of antibiotics as veterinary feed additives is potentially contributing to a significant reservoir of antibiotic resistance in agricultural farmlands via the application of antibiotic-contaminated manure. Vermicomposting of swine manure using housefly larvae is a promising biotechnology for waste reduction and control of antibiotic pollution. To determine how vermicomposting influences antibiotic resistance traits in swine manure, we explored the resistome and associated bacterial community dynamics during larvae gut transit over 6 days of treatment. In total, 94 out of 158 antibiotic resistance genes (ARGs) were significantly attenuated (by 85%), while 23 were significantly enriched (3.9-fold) following vermicomposting. The manure-borne bacterial community showed a decrease in the relative abundance of Bacteroidetes, and an increase in Proteobacteria, specifically Ignatzschineria, following gut transit. ARG attenuation was significantly correlated with changes in microbial community succession, especially reduction in Clostridiales and Bacteroidales. Six genomes were assembled from the manure, vermicompost (final product) and gut samples, including Pseudomonas, Providencia, Enterococcus, Bacteroides and Alcanivorax. Transposon-linked ARGs were more abundant in gut-associated bacteria compared with those from manure and vermicompost. Further, ARG-transposon gene cassettes had a high degree of synteny between metagenomic assemblies from gut and vermicompost samples, highlighting the significant contribution of gut microbiota through horizontal gene transfer to the resistome of vermicompost. In conclusion, the larvae gut microbiome significantly influences manure-borne community succession and the antibiotic resistome during animal manure processing.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Alcock RE, Sweetman A, Jones KC . (1999). Assessment of organic contaminant fate in waste water treatment plants I: Selected compounds and physicochemical properties. Chemosphere 38: 2247–2262.
Aminov RI . (2011). Horizontal gene exchange in environmental microbiota. Front Microbiol 2: 158.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9: 75.
Bernard L, Chapuis-Lardy L, Razafimbelo T, Razafindrakoto M, Pablo AL, Legname E et al. (2012). Endogeic earthworms shape bacterial functional communities and affect organic matter mineralization in a tropical soil. ISME J 6: 213–222.
Binh CTT, Heuer H, Kaupenjohann M, Smalla K . (2009). Diverse aadA gene cassettes on class 1 integrons introduced into soil via spread manure. Res Microbiol 160: 427–433.
Blahovec J, Kostecka Z, Kocisova A . (2006). Peptidolytic enzymes in different larval stadium of housefly Musca domestica. Vet Med-Czech 51: 139–144.
Boucher Y, Labbate M, Koenig JE, Stokes HW . (2007). Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol 15: 301–309.
Brinkmann N, Tebbe CC . (2007). Leaf-feeding larvae of Manduca sexta (Insecta, Lepidoptera) drastically reduce copy numbers of aadA antibiotic resistance genes from transplastomic tobacco but maintain intact aadA genes in their feces. Environ Biosafety Res 6: 121–133.
Calvert C, Morgan N, Martin R . (1970). House fly larvae: biodegradation of hen excreta to useful products. Poultry Sci 49: 588–589.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145.
Dominguez J, Aira M, Gomez-Brandon M . (2010). Vermicomposting: earthworms enhance the work of microbes. In: Insam H, Whittle IF, Goberna M (eds) Microbes at Work. Springer-Verlag: Berlin, Heidelberg, 93–114.
Egert M, Wagner B, Lemke T, Brune A, Friedrich MW . (2003). Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl Environ Microbiol 69: 6659–6668.
Egert M, Marhan S, Wagner B, Scheu S, Friedrich MW . (2004). Molecular profiling of 16S rRNA genes reveals diet-related differences of microbial communities in soil, gut, and casts of Lumbricus terrestris L. (Oligochaeta: Lumbricidae). FEMS Microbiol Ecol 48: 187–197.
Enne VI, Livermore DM, Stephens P, Hall LMC . (2001). Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357: 1325–1328.
Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG et al. (2013). Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4: 1111–1119.
Forsberg KJ, Reyes A, Bin W, Selleck EM, Sommer MOA, Dantas G . (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337: 1107–1111.
Gaze WH, Zhang LH, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J et al. (2011). Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J 5: 1253–1261.
Ghosh S, LaPara TM . (2007). The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J 1: 191–203.
Hartenstein R, Hartenstein F . (1981). Physicochemical changes effected in activated sludge by the earthworm Eisenia foetida. J Environ Qual 10: 377–381.
Heck K, De Marco EG, Duarte MW, Salamoni SP, Van der Sand S . (2015). Pattern of multiresistant to antimicrobials and heavy metal tolerance in bacteria isolated from sewage sludge samples from a composting process at a recycling plant in southern Brazil. Environ Monit Assess 187: 328.
Hvistendahl M . (2012). Public health China takes aim at rampant antibiotic resistance. Science 336: 795–795.
Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG et al. (2011). Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8: 761–U107.
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821.
Lemos FJA, Terra WR . (1991). Digestion of bacteria and the role of midgut lysozyme in some insect larvae. Comp Biochem Phys B 100: 265–268.
Li B, Yang Y, Ma LP, Ju F, Guo F, Tiedje JM et al. (2015a). Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J 9: 2490–2502.
Li DH, Liu CM, Luo RB, Sadakane K, Lam TW . (2015b). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31: 1674–1676.
Li H, Durbin R . (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760.
Liu B, Pop M . (2009). ARDB-antibiotic resistance genes database. Nucleic Acids Res 37: D443–D447.
Liu QL, Tomberlin JK, Brady JA, Sanford MR, Yu ZN . (2008). Black soldier fly (Diptera: Stratiomyidae) larvae reduce Escherichia coli in dairy manure. Environ Entomol 37: 1525–1530.
Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD et al. (2012). In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA 109: 1691–1696.
Mayende L, Wilhelmi BS, Pletschke BI . (2006). Cellulases (CMCases) and polyphenol oxidases from thermophilic Bacillus spp. isolated from compost. Soil Biol Biochem 38: 2963–2966.
Mazel D . (2006). Integrons: agents of bacterial evolution. Nat Rev Microbiol 4: 608–620.
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618.
Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM . (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J 8: 1577–1587.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M . (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35: W182–W185.
Mumcuoglu KY, Miller J, Mumcuoglu M, Friger M, Tarshis M . (2001). Destruction of bacteria in the digestive tract of the maggot of Lucilia sericata (Diptera: Calliphoridae). J Med Entomol 38: 161–166.
Naas T, Mikami Y, Imai T, Poirel L, Nordmann P . (2001). Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J Bacteriol 183: 235–249.
Neuhauser E, Kaplan D, Malecki M, Hartenstein R . (1980). Materials supporting weight gain by the earthworm Eisenia foetida in waste conversion systems. Agricul Wastes 2: 43–60.
Ozogul F . (2004). Production of biogenic amines by Morganella morganii, Klebsiella pneumoniae and Hafnia alvei using a rapid HPLC method. Eur Food Res Technol 219: 465–469.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW . (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055.
Petridis M, Bagdasarian M, Waldor MK, Walker E . (2006). Horizontal transfer of shiga toxin and antibiotic resistance genes among Escherichia coli strains in house fly (Diptera: Muscidae) gut. J Med Entomol 43: 288–295.
Pfaffl MW . (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
Poole TL, Callaway TR, Bischoff KM, Warnes CE, Nisbet DJ . (2006). Macrolide inactivation gene cluster mphA-mrx-mphR adjacent to a class 1 integron in Aeromonas hydrophila isolated from a diarrhoeic pig in Oklahoma. J Antimicrob Chemoth 57: 31–38.
Ramsden S, Ghosh S, Bohl L, LaPara T . (2010). Phenotypic and genotypic analysis of bacteria isolated from three municipal wastewater treatment plants on tetracycline-amended and ciprofloxacin‐amended growth media. J Appl Microbiol 109: 1609–1618.
Rho MN, Tang HX, Ye YZ . (2010). FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 38: e191.
Riber L, Poulsen PHB, Al-Soud WA, Hansen LBS, Bergmark L, Brejnrod A et al. (2014). Exploring the immediate and long-term impact on bacterial communities in soil amended with animal and urban organic waste fertilizers using pyrosequencing and screening for horizontal transfer of antibiotic resistance. FEMS Microbiol Ecol 90: 206–224.
Seemann T . (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069.
Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ . (2011). Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480: 241–244.
Su JQ, Wei B, Xu CY, Qiao M, Zhu YG . (2014). Functional metagenomic characterization of antibiotic resistance genes in agricultural soils from China. Environ Interl 65: 9–15.
Su JQ, Wei B, Ou-Yang WY, Huang FY, Zhao Y, Xu HJ et al. (2015). Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ Sci Technol 49: 7356–7363.
Su Z, Zhang M, Liu X, Tong L, Huang Y, Li G et al. (2010). Comparison of bacterial diversity in wheat bran and in the gut of larvae and newly emerged adult of Musca domestica (Diptera: Muscidae) by use of ethidium monoazide reveals bacterial colonization. J Economic Entomol 103: 1832–1841.
Wall DP, Deluca T . (2007). Ortholog detection using the reciprocal smallest distance algorithm. Methods Mol Biol 396: 95–110.
Wang FH, Ma WQ, Dou ZX, Ma L, Liu XL, Xu JX et al. (2006). The estimation of the production amount of animal manure and its environmental effect in China. China Environ Sci 26: 614–617.
Wang FH, Qiao M, Su JQ, Chen Z, Zhou X, Zhu YG . (2014). High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environ Sci Technol 48: 9079–9085.
Wang H, Zhang Z, Czapar GF, Winkler MK, Zheng J . (2013). A full-scale house fly (Diptera: Muscidae) larvae bioconversion system for value-added swine manure reduction. Waste Manage Res 31: 223–231.
Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT et al. (2007). Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450: 560–U517.
Wood DE, Salzberg SL . (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15: R46.
Xia LC, Steele JA, Cram JA, Cardon ZG, Simmons SL, Vallino JJ et al. (2011). Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst Biol 5: S15.
Zhang Z, Wang H, Zhu J, Suneethi S, Zheng J . (2012). Swine manure vermicomposting via housefly larvae (Musca domestica): the dynamics of biochemical and microbial features. Bioresour Technol 118: 563–571.
Zhang ZJ, Shen JG, Wang H, Liu M, Wu LH, Ping F et al. (2014). Attenuation of veterinary antibiotics in full-scale vermicomposting of swine manure via the housefly larvae (Musca domestica). Sci Rep-Uk 4: 6844.
Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci USA 110: 3435–3440.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (41373074, 21210008), Zhejiang Science and Technology Innovation Program (2013C33001, 2015C03SA420001), and the National Key Research and Development Plan (2016YFD0900205). This work was supported in part by the US Department of Energy under Contract DE-AC02-06CH11357.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, H., Sangwan, N., Li, HY. et al. The antibiotic resistome of swine manure is significantly altered by association with the Musca domestica larvae gut microbiome. ISME J 11, 100–111 (2017). https://doi.org/10.1038/ismej.2016.103
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2016.103
This article is cited by
-
Valorization of soybean-processing wastewater sludge via black soldier fly larvae: insights into the performance and bacterial community dynamics
Frontiers of Environmental Science & Engineering (2025)
-
Microplastics as vectors for antibiotic resistance genes and their implications for gut health
Discover Medicine (2025)
-
Organic fertilization co-selects genetically linked antibiotic and metal(loid) resistance genes in global soil microbiome
Nature Communications (2024)
-
Effects of magnesium-modified biochar on antibiotic resistance genes and microbial communities in chicken manure composting
Environmental Science and Pollution Research (2023)
-
Prevalence of macrolide–lincosamide–streptogramin resistant lactic acid bacteria isolated from food samples
Journal of Food Science and Technology (2023)