Abstract
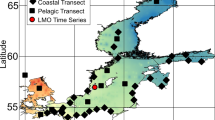
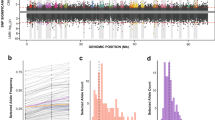
Members of the marine genus Rhodopirellula are attached living bacteria and studies based on cultured Rhodopirellula strains suggested that three closely related species R. baltica, ‘R. europaea’ and ‘R. islandica’ have a limited geographic distribution in Europe. To address this hypothesis, we developed a nested PCR for a single gene copy detection of a partial acetyl CoA synthetase (acsA) from intertidal sediments collected all around Europe. Furthermore, we performed growth experiments in a range of temperature, salinity and light conditions. A combination of Basic Local Alignment Search Tool (BLAST) and Minimum Entropy Decomposition (MED) was used to analyze the sequences with the aim to explore the geographical distribution of the species and subspecies. MED has been mainly used for the analysis of the 16S rRNA gene and here we propose a protocol for the analysis of protein-coding genes taking into account the degeneracy of the codons and a possible overestimation of functional diversity. The high-resolution analysis revealed differences in the intraspecies community structure in different geographic regions. However, we found all three species present in all regions sampled and in agreement with growth experiments we demonstrated that Rhodopirellula species do not have a limited geographic distribution in Europe.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bengtsson MM, Øvreås L . (2010). Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol 10: 261.
Barberán A, Ladau J, Leff JW, Pollard KS, Menninger HL, Dunn RR, Fierer N . (2015). Continental-scale distributions of dust-associated bacteria and fungi. Proc Natl Acad Sci USA 112: 5756–5761.
Bondoso J, Albuquerque L, Lobo-da-Cunha A, Da Costa MS, Harder J, Lage OM . (2014). Rhodopirellula lusitana sp. nov. and Rhodopirellula rubra sp. nov., isolated from the surface of macroalgae. Syst Appl Microbiol 37: 157–164.
Bondoso J, Harder J, Lage OM . (2013). rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Van Leeuwenhoek 104: 477–488.
Brown SDJ, Collins RA, Boyer S, Lefort M-C, Malumbres-Olarte J, Vink CJ et al. (2012). Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol Ecol Resour 12: 562–565.
Buttigieg PL, Ramette A . (2015). Biogeographic patterns of bacterial microdiversity in Arctic deep-sea sediments (HAUSGARTEN, Fram Strait). Front Microbiol 5: 660.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 421–421.
Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA . (2012). The Western English Channel contains a persistent microbial seed bank. ISME J 6: 1089–1093.
Chao A, Gotelli NJ, Hsieh TC, Sander EL, Ma KH, Colwell RK et al. (2014). Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monogr 84: 45–67.
DeLong EF, Franks DG, Alldredge AL . (1993). Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr 38: 924–934.
Eren AM, Borisy GG, Huse SM, Mark Welch JL . (2014a). Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA 111: E2875–E2884.
Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG et al. (2013). Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4: 1111–1119.
Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML . (2014b). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9: 968–979.
Eren AM, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ et al. (2015). A single genus in the gut microbiome reflects host preference and specificity. ISME J 9: 90–100.
Fuerst JA, Gwilliam HG, Lindsay M, Lichanska A, Belcher C, Vickers JE et al. (1997). Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn Penaeus monodon. Appl Environ Microbiol 63: 254–262.
Fuerst JA, Sagulenko E . (2011). Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9: 403–413.
Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W et al. (2003). Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci 100: 8298–8303.
Gouy M, Guindon S, Gascuel O . (2009). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224.
Katoh K, Standley DM . (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780.
Kimura M . (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120.
Kizina J, Žure M, Rhiel E, Munn CB, Richter M, Harder J . (2015). Permanent draft genome of ‘Rhodopirellula islandica’ strain K833. Mar Genomics 24: 249–251.
Li W, Godzik A . (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659.
Martin M . (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10+.
McLellan SL, Newton RJ, Vandewalle JL, Shanks OC, Huse SM, Eren AM et al. (2013). Sewage reflects the distribution of human faecal Lachnospiraceae. Environ Microbiol 15: 2213–2227.
Paradis E, Claude J, Strimmer K . (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290.
Paulson JN, Stine OC, Bravo HC, Pop M . (2013). Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10: 1200–1202.
Pimentel-Elardo S, Wehrl M, Friedrich AB, Jensen PR, Hentschel U . (2003). Isolation of planctomycetes from Aplysina sponges. Aquat Microb Ecol 33: 239–245.
Ramette A, Tiedje JM . (2007). Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol 53: 197–207.
Rice P, Longden I, Bleasby A . (2000). EMBOSS: the european molecular biology open software suite. Trends Genet 16: 276–277.
Richter-Heitmann T, Richter M, Klindworth A, Wegner C-E-E, Frank CS, Glöckner FO et al. (2014). Permanent draft genomes of the two Rhodopirellula europaea strains 6C and SH398. Mar Genomics 13: 15–16.
Richter M, Richter-Heitmann T, Klindworth A, Wegner C-E-E, Frank CS, Harder J et al. (2014). Permanent draft genomes of the three Rhodopirellula baltica strains SH28, SWK14 and WH47. Mar Genomics 13: 13–14.
Roh SW, Lee H-W-W, Yim KJ, Shin N-R-R, Lee J, Whon TW et al. (2013). Rhodopirellula rosea sp. nov., a novel bacterium isolated from an ark clam Scapharca broughtonii. J Microbiol 51: 301–304.
Schlesner H . (1994). The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst Appl Microbiol 17: 135–145.
Schlesner H, Rensmann C, Tindall BJ, Gade D, Rabus R, Pfeiffer S et al. (2004). Taxonomic heterogeneity within the Planctomycetales as derived by DNA–DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol 54: 1567–1580.
Shannon CE . (1948). A mathematical theory of communication. Bell Syst Tech J 27: 379–423.
Sintes E, De Corte D, Haberleitner E, Herndl GJ . (2016). Geographic distribution of archaeal ammonia oxidizing ecotypes in the Atlantic Ocean. Front Microbiol 7: 77.
Speth DR, van Teeseling MC, Jetten MS . (2012). Genomic analysis indicates the presence of an asymmetric bilayer outer membrane in Planctomycetes and Verrucomicrobia. Front Microbiol 3: 304.
Strous M, Kuenen JG, Fuerst JA, Wagner M, Jetten MSM . (2002). The anammox case—a new experimental manifesto for microbiological eco-physiology. Antonie Van Leeuwenhoek 81: 693–702.
Wagner M, Horn M . (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17: 241–249.
Wecker P, Klockow C, Ellrott A, Quast C, Langhammer P, Harder J, Glöckner FO . (2009). Transcriptional response of the model planctomycete Rhodopirellula baltica SH1T to changing environmental conditions. BMC Genomics 10: 410.
Winkelmann N, Harder J . (2009). An improved isolation method for attached-living Planctomycetes of the genus Rhodopirellula. J Microbiol Methods 77: 276–284.
Winkelmann N, Jaekel U, Meyer C, Serrano W, Rachel R, Rosselló-Mora R et al. (2010). Determination of the diversity of Rhodopirellula isolates from European seas by multilocus sequence analysis. Appl Environ Microbiol 76: 776–785.
Yoon J, Matsuo Y, Kasai H, Lee M-K . (2015). Phylogenetic and taxonomic analyses of Rhodopirellula caenicola sp. nov., a new marine Planctomycetes species isolated from Iron Sand. J Phylogenet Evol Biol 3: 143.
Zhang J, Kobert K, Flouri T, Stamatakis A . (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30: 614–620.
Žure M, Munn CB, Harder J . (2015). Diversity of Rhodopirellula and related planctomycetes in a North Sea coastal sediment employing carB as molecular marker. FEMS Microbiol Lett 362: fnv127.
Acknowledgements
We would like to thank all sample contributors from EMBC (http://www.embcplus.org), MARES (http://www.mares-eu.org), Plymouth University and Max Planck-Institute, Bremen. We thank Maryia Khomich and Christina Probian for technical assistance. This work was supported by the Max Planck Society and MARES (Erasmus Mundus Joint Doctorate programme coordinated by Ghent University, FPA 2011-0016). AFG was supported by the Micro B3 project, which is funded from the European Union's Seventh Framework Programme (FP7; Joint Call OCEAN.2011-2: Marine microbial diversity – new insights into marine ecosystems functioning and its biotechnological potential) under grant agreement no. 287589.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Žure, M., Fernandez-Guerra, A., Munn, C. et al. Geographic distribution at subspecies resolution level: closely related Rhodopirellula species in European coastal sediments. ISME J 11, 478–489 (2017). https://doi.org/10.1038/ismej.2016.123
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2016.123