Abstract
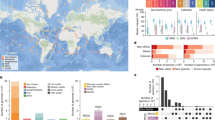
Marine Euryarchaeota remain among the least understood major components of marine microbial communities. Marine group II Euryarchaeota (MG-II) are more abundant in surface waters (4–20% of the total prokaryotic community), whereas marine group III Euryarchaeota (MG-III) are generally considered low-abundance members of deep mesopelagic and bathypelagic communities. Using genome assembly from direct metagenome reads and metagenomic fosmid clones, we have identified six novel MG-III genome sequence bins from the photic zone (Epi1–6) and two novel bins from deep-sea samples (Bathy1–2). Genome completeness in those genome bins varies from 44% to 85%. Photic-zone MG-III bins corresponded to novel groups with no similarity, and significantly lower GC content, when compared with previously described deep-MG-III genome bins. As found in many other epipelagic microorganisms, photic-zone MG-III bins contained numerous photolyase and rhodopsin genes, as well as genes for peptide and lipid uptake and degradation, suggesting a photoheterotrophic lifestyle. Phylogenetic analysis of these photolyases and rhodopsins as well as their genomic context suggests that these genes are of bacterial origin, supporting the hypothesis of an MG-III ancestor that lived in the dark ocean. Epipelagic MG-III occur sporadically and in relatively small proportions in marine plankton, representing only up to 0.6% of the total microbial community reads in metagenomes. None of the reconstructed epipelagic MG-III genomes were present in metagenomes from aphotic zone depths or from high latitude regions. Most low-GC bins were highly enriched at the deep chlorophyll maximum zones, with the exception of Epi1, which appeared evenly distributed throughout the photic zone worldwide.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH . (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538.
Alonso-Saez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL et al. (2012). Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA 109: 17989–17994.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Allers E, Wright JJ, Konwar KM, Howes CG, Beneze E, Hallam SJ et al. (2013). Diversity and population structure of Marine Group A bacteria in the Northeast subarctic Pacific Ocean. Isme J 7: 256–268.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477.
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S et al. (2004). The Pfam protein families database. Nucleic Acids Res 32 (Dataabase issue): D138–D141.
Batut B, Knibbe C, Marais G, Daubin V . (2014). Reductive genome evolution at both ends of the bacterial population size spectrum. Nat Rev Microbiol 12: 841–850.
Beja O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP et al. (2000). Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol 2: 516–529.
Belmar L, Molina V, Ulloa O . (2011). Abundance and phylogenetic identity of archaeoplankton in the permanent oxygen minimum zone of the eastern tropical South Pacific. FEMS Microbiol Ecol 78: 314–326.
Boutrif M, Garel M, Cottrell MT, Tamburini C . (2011). Assimilation of marine extracellular polymeric substances by deep-sea prokaryotes in the NW Mediterranean Sea. Environ Microbiol Rep 3: 705–709.
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P . (2008). Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6: 245–252.
Bryant JA, Aylward FO, Eppley JM, Karl DM, Church MJ, DeLong EF . (2015). Wind and sunlight shape microbial diversity in surface waters of the North Pacific Subtropical Gyre. ISME J 10: 1308–1322.
Cashmore AR, Jarillo JA, Wu YJ, Liu D . (1999). Cryptochromes: blue light receptors for plants and animals. Science 284: 760–765.
Coleman ML, Chisholm SW . (2010). Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA 107: 18634–18639.
Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF et al. (2006). Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770.
Cuadros-Orellana S, Martin-Cuadrado AB, Legault B, D'Auria G, Zhaxybayeva O, Papke RT et al. (2007). Genomic plasticity in prokaryotes: the case of the square haloarchaeon. ISME J 1: 235–245.
Chang YG, Cohen SE, Phong C, Myers WK, Kim YI, Tseng R et al. (2015). Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science 349: 324–328.
Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K et al. (2011). The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–364.
Chen S, Su L, Chen J, Wu J . (1997). Cutinase: characteristics, preparation, and application. Biotechnol Adv 31: 1754–1767.
DeLong EF . (1992). Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685–5689.
DeLong EF . (2006). Archaeal mysteries of the deep revealed. Proc Natl Acad Sci U S A 103: 6417–6418.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503.
DeLong EF, Taylor LT, Marsh TL, Preston CM . (1999). Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol 65: 5554–5563.
Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, López-García P . (2014). Pangenome evidence for extensive inter-domain horizontal transfer affecting lineage-core and shell genes in uncultured planktonic Thaumarchaeota and Euryarchaeota. Genome Biol Evol 6: 1549–1563.
Dufresne A, Garczarek L, Partensky F . (2005). Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol 6: R14.
Edgar RC . (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Edgcomb VP, Kysela DT, Teske A, de Vera Gomez A, Sogin ML . (2002). Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci USA 99: 7658–7662.
Essen LO . (2006). Photolyases and cryptochromes: common mechanisms of DNA repair and light-driven signaling? Curr Opin Struct Biol 16: 51–59.
Essen LO, Klar T . (2006). Light-driven DNA repair by photolyases. Cell Mol Life Sci 63: 1266–1277.
Fuhrman JA, Davis AA . (1997). Widespread archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. March Ecol Prog Series 150: 275–285.
Fuhrman JA, McCallum K, Davis AA . (1992). Novel major archaebacterial group from marine plankton. Nature 356: 148–149.
Fuhrman JA, Schwalbach MS, Stingl U . (2008). Proteorhodopsins: an array of physiological roles? Nat Rev Microbiol 6: 488–494.
Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C . (2009). Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J 3: 860–869.
Galand PE, Gutiérrez-Provecho C, Massana R, Gasol JM, Casamayor EO . (2010). Inter-annual recurrence of archaeal assemblages in the coastal NW Mediterranean Sea (Blanes Bay Microbial Observatory). Limnol Oceanogr. 55: 2117–2125.
Ghai R, Martin-Cuadrado A, Gonzaga A, Garcia-Heredia I, Cabrera R, Martin J et al. (2010). Metagenome of the Mediterranenan deep Chlorophyl Maximum stuidied by direct and fomid library 454 pyrosequencing. ISME J 4: 1154–1166.
Ghai R, McMahon KD, Rodriguez-Valera F . (2012). Breaking a paradigm: cosmopolitan and abundant freshwater actinobacteria are low GC. Environ Microbiol Rep 4: 29–35.
Giovannoni S, Nemergut D . (2014). Ecology. Microbes ride the current. Science 345: 1246–1247.
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM . (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91.
Gribaldo S, Lumia V, Creti R, Conway de Macario E, Sanangelantoni A, Cammarano P . (1999). Discontinuous occurrence of the hsp70 (dnaK) gene among Archaea and sequence features of HSP70 suggest a novel outlook on phylogenies inferred from this protein. J Bacteriol 181: 434–443.
Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT et al. (2001). TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res 29: 41–43.
Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J et al. (2006). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA 103: 18296–18301.
Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A et al. (2005). Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol 71: 2303–2309.
Huang Y, Gilna P, Li W . (2009). Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25: 1338–1340.
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ . (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119.
Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K et al. (2013). A light-driven sodium ion pump in marine bacteria. Nat Commun 4: 1678.
Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV . (2012). Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335: 587–590.
Jarrell KF, Jones GM, Kandiba L, Nair DB, Eichler J . (2010) S-layer glycoproteins and flagellins: reporters of archaeal posttranslational modifications. Archaea 2010.
Jarrell KF, McBride MJ . (2008). The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6: 466–476.
Karner MB, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510.
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Konstantinidis KT, Tiedje JM . (2005). Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102: 2567–2572.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645.
Larsson J, Celepli N, Ininbergs K, Dupont CL, Yooseph S, Bergman B et al. (2014). Picocyanobacteria containing a novel pigment gene cluster dominate the brackish water Baltic Sea. ISME J.
Lassak K, Ghosh A, Albers SV . (2012). Diversity, assembly and regulation of archaeal type IV pili-like and non-type-IV pili-like surface structures. Res Microbiol 163: 630–644.
Lassmann T, Sonnhammer EL . (2005). Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6: 298.
Li H, Durbin R . (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760.
Li M, Baker BJ, Anantharaman K, Jain S, Breier JA, Dick GJ . (2015). Genomic and transcriptomic evidence for scavenging of diverse organic compounds by widespread deep-sea archaea. Nat Commun 6: 8933.
Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F et al. (2015). Ocean plankton. Determinants of community structure in the global plankton interactome. Science 348: 1262073.
Lopez-Garcia P, Lopez-Lopez A, Moreira D, Rodriguez-Valera F . (2001a). Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microbiol Ecol 36: 193–202.
Lopez-Garcia P, Moreira D, Lopez-Lopez A, Rodriguez-Valera F . (2001b). A novel haloarchaeal-related lineage is widely distributed in deep oceanic regions. Environ Microbiol 3: 72–78.
Lopez-Garcia P, Philippe H, Gail F, Moreira D . (2003). Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc Natl Acad Sci USA 100: 697–702.
Lopez-Garcia P, Zivanovic Y, Deschamps P, Moreira D . (2016). Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol 13: 447–456.
Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI et al. (1999). Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res 9: 608–628.
Makarova KS, Koonin EV . (2003). Comparative genomics of Archaea: how much have we learned in six years, and what's next? Genome Biol 4: 115.
Makarova KS, Wolf YI, Koonin EV . (2015) archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales Life (Basel) 5: 818–840.
Martin-Cuadrado AB, Fontaine T, Esteban PF, del Dedo JE, de Medina-Redondo M, del Rey F et al. (2008). Characterization of the endo-beta-1,3-glucanase activity of S. cerevisiae Eng2 and other members of the GH81 family. Fungal Genet Biol 45: 542–553.
Martin-Cuadrado AB, Garcia-Heredia I, Molto AG, Lopez-Ubeda R, Kimes N, Lopez-Garcia P et al. (2015). A new class of marine Euryarchaeota group II from the Mediterranean deep chlorophyll maximum. ISME J 9: 1619–1634.
Martin-Cuadrado AB, Pasic L, Rodriguez-Valera F . (2014). Diversity of the cell-wall associated genomic island of the archaeon Haloquadratum walsbyi. BMC Genomics 16: 603.
Martin-Cuadrado AB, Rodriguez-Valera F, Moreira D, Alba JC, Ivars-Martinez E, Henn MR et al. (2008). Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J 2: 865–886.
Massana R, DeLong EF, Pedros-Alio C . (2000). A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl Environ Microbiol 66: 1777–1787.
Mizuno CM, Ghai R, Saghai A, Lopez-Garcia P, Rodriguez-Valera F . (2016). Genomes of abundant and widespread viruses from the deep ocean. MBio 7: 4.
Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ et al. (2012). De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6: 81–93.
Nawrocki EP . (2009) Structural RNA homology search and alignment using covariance models, PhD thesis. Washington University in Saint Louis, School of Medicine, St. Louis, MO, USA.
Needham DM, Fuhrman JA . (2016). Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol.
Orsi WD, Smith JM, Wilcox HM, Swalwell JE, Carini P, Worden AZ et al. (2015). Ecophysiology of uncultivated marine euryarchaea is linked to particulate organic matter. ISME J 9: 1747–1763.
Ouverney CC, Fuhrman JA . (2000). Marine planktonic archaea take up amino acids. Appl Environ Microbiol 66: 4829–4833.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW . (2014). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055.
Peng Y, Leung HC, Yiu SM, Chin FY . (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428.
Petitjean C, Moreira D, Lopez-Garcia P, Brochier-Armanet C . (2012). Horizontal gene transfer of a chloroplast DnaJ-Fer protein to Thaumarchaeota and the evolutionary history of the DnaK chaperone system in Archaea. BMC Evol Biol 12: 226.
Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA 111: 12504–12509.
Quaiser A, Zivanovic Y, Moreira D, Lopez-Garcia P . (2011). Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5: 285–304.
Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P . (2007). Prediction of effective genome size in metagenomic samples. Genome Biol 8: R10.
Raymann K, Forterre P, Brochier-Armanet C, Gribaldo S . (2014). Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in archaea. Genome Biol Evol 6: 192–212.
Reva O, Tummler B . (2008). Think big—giant genes in bacteria. Environ Microbiol 10: 768–777.
Rice P, Longden I, Bleasby A . (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277.
Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B et al. (2009). Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7: 828–836.
Romero H, Pereira E, Naya H, Musto H . (2009). Oxygen and guanine-cytosine profiles in marine environments. J Mol Evol 69: 203–206.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5: e77.
Scheerer P, Zhang F, Kalms J, von Stetten D, Krauss N, Oberpichler I et al. (2015). The class III cyclobutane pyrimidine dimer photolyase structure reveals a new antenna chromophore binding site and alternative photoreduction pathways. J Biol Chem 290: 11504–11514.
Schwarz G, Mendel RR, Ribbe MW . (2009). Molybdenum cofactors, enzymes and pathways. Nature 460: 839–847.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504.
Smedile F, Messina E, La Cono V, Tsoy O, Monticelli LS, Borghini M et al. (2012). Metagenomic analysis of hadopelagic microbial assemblages thriving at the deepest part of Mediterranean Sea, Matapan-Vavilov Deep. Environ Microbiol 3: 1462–2920.
Strom SL, Brahamsha B, Fredrickson KA, Apple JK, Rodriguez AG . (2011). A giant cell surface protein in Synechococcus WH8102 inhibits feeding by a dinoflagellate predator. Environ Microbiol.
Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G et al. (2015). Ocean plankton. Structure and function of the global ocean microbiome. Science 348: 1261359.
Swan BK, Chaffin MD, Martinez-Garcia M, Morrison HG, Field EK, Poulton NJ et al. (2014). Genomic and metabolic diversity of Marine Group I Thaumarchaeota in the mesopelagic of two subtropical gyres. PLoS One 9: e95380.
Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, Gonzalez JM et al. (2013). Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA 110: 11463–11468.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S . (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729.
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43.
Vavourakis CD, Ghai R, Rodriguez-Valera F, Sorokin DY, Tringe SG, Hugenholtz P et al. (2016). Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline Soda Lake Brines. Front Microbiol 7: 211.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Acknowledgements
We are thankful to José de la Torre for editing the manuscript. We are thankful to L Gasperini and G Bortoluzzi of the Instituto di Geologia Marina (ISMAR), CNR, Bologna (Italy) for allowing PLG to participate in the Marmara2010 R/V Urania cruise during which part of the samples analyzed in this study were collected. This work was supported by projects MEDIMAX BFPU2013–48007-P from the Spanish Ministerio de Economía y Competitividad, MaCuMBA Project 311975 of the European Commission FP7, project AQUAMET II/2014/012 from the Generalitat Valenciana and by the French Agence Nationale de la Recherche (ANR-08-GENM-024–001,EVOLDEEP). JHM was supported with a PhD fellowship from the Spanish Ministerio de Economía y Competitividad.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Haro-Moreno, J., Rodriguez-Valera, F., López-García, P. et al. New insights into marine group III Euryarchaeota, from dark to light. ISME J 11, 1102–1117 (2017). https://doi.org/10.1038/ismej.2016.188
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2016.188
This article is cited by
-
Depth-dependent microbial metagenomes sampled in the northeastern Indian Ocean
Scientific Data (2024)
-
Insights into the prokaryotic communities of the abyssal-hadal benthic-boundary layer of the Kuril Kamchatka Trench
Environmental Microbiome (2023)
-
Picoplankton diversity in an oligotrophic and high salinity environment in the central Adriatic Sea
Scientific Reports (2023)
-
Virus diversity and interactions with hosts in deep-sea hydrothermal vents
Microbiome (2022)
-
Identification of a protein responsible for the synthesis of archaeal membrane-spanning GDGT lipids
Nature Communications (2022)