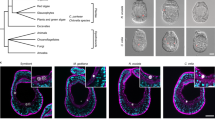
Abstract
Analysis of the widely used ITS region is confounded by the presence of intragenomic variants (IGVs). In Symbiodinium, the algal symbionts of reef building corals, deep-sequencing analyses are used to characterise communities within corals, yet these analyses largely overlook IGVs. Here we consider that distinct ITS2 sequences could represent IGVs rather than distinct symbiont types and argue that symbionts can be distinguished by their proportional composition of IGVs, described as their ITS2 metahaplotype. Using our metahaplotype approach on Minimum Entropy Decomposition (MED) analysis of ITS2 sequences from the corals Acropora downingi, Cyphastrea microphthalma and Playgyra daedalea, we show the dominance of a single species-specific Symbiodinium C3 variant within each coral species. We confirm the presence of these species-specific symbionts using the psbA non-coding region. Our findings highlight the importance of accounting for IGVs in ITS2 analyses and demonstrate their capacity to resolve biological patterns that would otherwise be overlooked.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Álvarez I, Wendel JF . (2003). Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol 29: 417–434.
Arif C, Daniels C, Bayer T, Banguera‐Hinestroza E, Barbrook A, Howe CJ et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next‐generation sequencing‐based genotyping of the ITS2 rDNA region. Mol Ecol 23: 4418–4433.
Berkelmans R, Van Oppen MJ . (2006). The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc R Soc B Biol Sci 273: 2305–2312.
Cunning R, Yost DM, Guarinello ML, Putnam HM, Gates RD . (2015). Variability of Symbiodinium communities in waters, sediments, and corals of thermally distinct reef pools in American Samoa. PLoS One 10: e0145099.
Eren AM, Borisy GG, Huse SM, Welch JLM . (2014). Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA 111: E2875–E2884.
Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML . (2015). Minimum entropy decomposition: Unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9: 968–979.
Hume B, D'Angelo C, Smith E, Stevens J, Burt J, Wiedenmann J . (2015). Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world's hottest sea, the Persian/Arabian Gulf. Sci Rep 5: 8562.
LaJeunesse TC . (2001). Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: in search of a ‘species’ level marker. J Phycol 37: 866–880.
LaJeunesse TC . (2005). ‘Species’ radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition. Mol Biol Evol 22: 570–581.
LaJeunesse TC, Thornhill DJ . (2011). Improved resolution of reef-coral endosymbiont (Symbiodinium) species diversity, ecology, and evolution through psbA non-coding region genotyping. PLoS ONE 6: e29013.
Mai JC, Coleman AW . (1997). The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J Mol Evol 44: 258–271.
Muscatine L, Falkowski P, Porter J, Dubinsky Z . (1984). Fate of photosynthetic fixed carbon in light-and shade-adapted colonies of the symbiotic coral Stylophora pistillata. Proc R Soc Lond B Biol Sci 222: 181–202.
Sampayo E, Franceschinis L, Hoegh-Guldberg O, Dove S . (2007). Niche partitioning of closely related symbiotic dinoflagellates. Mol Ecol 16: 3721.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109: 6241–6246.
Stat M, Bird CE, Pochon X, Chasqui L, Chauka LJ, Concepcion GT et al. (2011). Variation in Symbiodinium ITS2 sequence assemblages among coral colonies. PLoS ONE 6: e15854.
Stern RF, Andersen RA, Jameson I, Küpper FC, Coffroth M-A, Vaulot D et al. (2012). Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS ONE 7: e42780.
Thornhill DJ, Lajeunesse TC, Santos SR . (2007). Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol Ecol 16: 5326–5340.
Thornhill DJ, Lewis AM, Wham DC, LaJeunesse TC . (2014). Host‐specialist lineages dominate the adaptive radiation of reef coral endosymbionts. Evolution 68: 352–367.
Acknowledgements
We thank the New York University Abu Dhabi Institute for funding and the Environment Agency Abu Dhabi for permitting. This work was supported by the New York University Core Sequencing and Bioinformatics Groups. Data used in this study are deposited in the Dryad Digital Repository. (http://dx.doi.org/10.5061/dryad.h6s54).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on ISME J website
Supplementary information
Rights and permissions
About this article
Cite this article
Smith, E., Ketchum, R. & Burt, J. Host specificity of Symbiodinium variants revealed by an ITS2 metahaplotype approach. ISME J 11, 1500–1503 (2017). https://doi.org/10.1038/ismej.2016.206
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2016.206
This article is cited by
-
Molecular identification and studies on genetic diversity and structure-related GC heterogeneity of Spatholobus Suberectus based on ITS2
Scientific Reports (2024)
-
Micro and macroevolution of sea anemone venom phenotype
Nature Communications (2023)
-
Mutualistic dinoflagellates with big disparities in ribosomal DNA variation may confound estimates of symbiont diversity and ecology in the jellyfish Cotylorhiza tuberculata
Symbiosis (2022)
-
Global-Scale Diversity and Distribution Characteristics of Reef-Associated Symbiodiniaceae via the Cluster-Based Parsimony of Internal Transcribed Spacer 2 Sequences
Journal of Ocean University of China (2021)
-
Coral microbiome composition along the northern Red Sea suggests high plasticity of bacterial and specificity of endosymbiotic dinoflagellate communities
Microbiome (2020)