Abstract
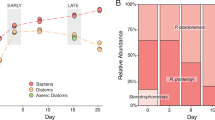
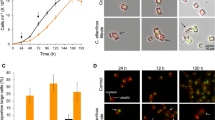
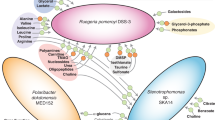
In their role as primary producers, marine phytoplankton modulate heterotrophic bacterial activities through differences in the types and amounts of organic matter they release. This study investigates the transcriptional response of bacterium Ruegeria pomeroyi, a member of the Roseobacter clade known to affiliate with diverse phytoplankton groups in the ocean, during a shift in phytoplankton taxonomy. The bacterium was initially introduced into a culture of the dinoflagellate Alexandrium tamarense, and then experienced a change in phytoplankton community composition as the diatom Thalassiosira pseudonana gradually outcompeted the dinoflagellate. Samples were taken throughout the 30-day experiment to track shifts in bacterial gene expression informative of metabolic and ecological interactions. Transcriptome data indicate fundamental differences in the exometabolites released by the two phytoplankton. During growth with the dinoflagellate, gene expression patterns indicated that the main sources of carbon and energy for R. pomeroyi were dimethysulfoniopropionate (DMSP), taurine, methylated amines, and polyamines. During growth with the diatom, dihydroxypropanesulfonate (DHPS), xylose, ectoine, and glycolate instead appeared to fuel the bulk of bacterial metabolism. Expression patterns of genes for quorum sensing, gene transfer agent, and motility suggest that bacterial processes related to cell communication and signaling differed depending on which phytoplankton species dominated the co-culture. A remodeling of the R. pomeroyi transcriptome implicating more than a quarter of the genome occurred through the change in phytoplankton regime.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101.
Amin SA, Parker MS, Armbrust EV . (2012). Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76: 667–684.
Anders S, Pyl PT, Huber W . (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169.
Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86.
Azam F, Fenchel T, Field J, Gray J, Meyer-Reil L, Thingstad F . (1983). The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10: 257–263.
Bathmann UV, Scharek R, Klaas C, Dubischar CD, Smetacek V . (1997). Spring development of phytoplankton biomass and composition in major water masses of the Atlantic sector of the Southern Ocean. Deep Sea Res Part II Top Stud Oceanogr 44: 51–67.
Becker JW, Berube PM, Follett CL, Waterbury JB, Chisholm SW, DeLong EF et al. (2014). Closely related phytoplankton species produce similar suites of dissolved organic matter. Front Microbiol 5: 111.
Biers EJ, Wang K, Pennington C, Belas R, Chen F, Moran MA . (2008). Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl Environ Microbiol 74: 2933–2939.
Biller SJ, Coe A, Chisholm SW . (2016). Torn apart and reunited: impact of a heterotroph on the transcriptome of Prochlorococcus. ISME J 10: 2831–2843.
Bürgmann H, Howard EC, Ye W, Sun F, Sun S, Napierala S et al. (2007). Transcriptional response of Silicibacter pomeroyi DSS-3 to dimethylsulfoniopropionate (DMSP). Environ Microbiol 9: 2742–2755.
Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC . (2011). Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci 108: 17791–17796.
Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG . (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93.
Cunliffe M . (2016). Purine catabolic pathway revealed by transcriptomics in the model marine bacterium Ruegeria pomeroyi DSS-3. FEMS Microbiol Ecol 92: 150.
Denger K, Mayer J, Buhmann M, Weinitschke S, Smits THM, Cook AM . (2009). Bifurcated degradative pathway of 3-sulfolactate in Roseovarius nubinhibens ISM via sulfoacetaldehyde acetyltransferase and (S)-cysteate sulfolyase. J Bacteriol 191: 5648–5656.
Denger K, Smits THM, Cook AM . (2006). L-Cysteate sulpho-lyase, a widespread pyridoxal 5′-phosphate-coupled desulphonative enzyme purified from Silicibacter pomeroyi DSS-3 T. Biochem J 394: 657–664.
Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ et al. (2015). Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci 112: 453–457.
Gerdes SY, Scholle MD, Campbell JW, Balázsi G, Ravasz E, Daugherty MD et al. (2003). Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 185: 5673–5684.
González JM, Covert JS, Whitman WB, Henriksen JR, Mayer F, Scharf B et al. (2003). Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int J Syst Evol Microbiol 53: 1261–1269.
González JM, Simó R, Massana R, Covert JS, Casamayor EO, Pedrós-Alió C et al. (2000). Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol 66: 4237–4246.
Gorzynska AK, Denger K, Cook AM, Smits THM . (2006). Inducible transcription of genes involved in taurine uptake and dissimilation by Silicibacter pomeroyi DSS-3 T. Arch Microbiol 185: 402.
Grossart H, Simon M . (2007). Interactions of planktonic algae and bacteria: effects on algal growth and organic matter dynamics. Aquat Microb Ecol 47: 163–176.
Hansell DA . (2013). Recalcitrant dissolved organic carbon fractions. Annu Rev Mar Sci 5: 421–445.
Harvey EL, Deering RW, Rowley DC, El Gamal A, Schorn M, Moore BS et al. (2016). A bacterial quorum-sensing precursor induces mortality in the marine coccolithophore, Emiliania huxleyi. Front Microbiol 7: 59.
Hasle G . (1976). The biogeography of some marine planktonic diatoms. Deep Sea Res Oceanogr Abstr 23: 319–338.
Hellebust JA . (1965). Excretion of some organic compounds by marine phytoplankton. Limnol Ocean 10: 192–206.
Howard EC, Henriksen JR, Buchan A, Reisch CR, Bürgmann H, Welsh R et al. (2006). Bacterial taxa that limit sulfur flux from the ocean. Science 314: 649–652.
Jasti S, Sieracki ME, Poulton NJ, Giewat MW, Rooney-Varga JN . (2005). Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl Environ Microbiol 71: 3483–3494.
Johnson WM, Kido Soule MC, Kujawinski EB . (2016). Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J 10: 2304–2316.
Keller MD . (1989). Dimethyl sulfide production and marine phytoplankton: the importance of species composition and cell size. Biol Oceanogr 6: 375–382.
Keller MD, Kiene RP, Matrai PA, Bellows WK . (1999). Production of glycine betaine and dimethylsulfoniopropionate in marine phytoplankton. I. Batch cultures. Mar Biol 135: 237–248.
Kohonen T . (2001) Self-organizing Maps. Springer: Germany.
Lafay B, Ruimy R, Rausch De Traubenberg C, Breittmayer V, Gauthier MJ, Christen R . (1995). Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int J Syst Evol Microbiol 45: 290–296.
Lang AS, Beatty JT . (2002). A bacterial signal transduction system controls genetic exchange and motility. J Bacteriol 184: 913–918.
Langmead B, Salzberg SL . (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359.
Lau WWY, Armbrust EV . (2006). Detection of glycolate oxidase gene glcD diversity among cultured and environmental marine bacteria. Environ Microbiol 8: 1688–1702.
Lee C, Jørgensen NOG . (1995). Seasonal cycling of putrescine and amino acids in relation to biological production in a stratified coastal salt pond. Biogeochemistry 29: 131–157.
Leung MM, Brimacombe CA, Spiegelman GB, Beatty JT . (2012). The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol Microbiol 83: 759–774.
Li MZ, Elledge SJ . (2007). Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4: 251–256.
Lidbury ID, Murrell JC, Chen Y . (2015). Trimethylamine and trimethylamine N-oxide are supplementary energy sources for a marine heterotrophic bacterium: implications for marine carbon and nitrogen cycling. ISME J 9: 760–769.
Love MI, Huber W, Anders S . (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550.
Luo H, Moran MA . (2014). Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78: 573–587.
Mayer J, Huhn T, Habeck M, Denger K, Hollemeyer K, Cook AM . (2010). 2,3-Dihydroxypropane-1-sulfonate degraded by Cupriavidus pinatubonensis JMP134: purification of dihydroxypropanesulfonate 3-dehydrogenase. Microbiology 156: 1556–1564.
Mercer RG, Quinlan M, Rose AR, Noll S, Beatty JT, Lang AS . (2012). Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol Lett 331: 53–62.
Miller MB, Bassler BL . (2001). Quorum sensing in bacteria. Annu Rev Microbiol 55: 165–199.
Miller TR, Belas R . (2006). Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ Microbiol 8: 1648–1659.
Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S et al. (2007). Ecological genomics of marine roseobacters. Appl Environ Microbiol 73: 4559–4569.
Moran MA, Buchan A, González JM, Heidelberg JF, Whitman WB, Kiene RP et al. (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432: 910–913.
Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwihare LI, Buchan A et al. (2016). Deciphering ocean carbon in a changing world. Proc Natl Acad Sci 113: 3143–3151.
Mou X, Sun S, Rayapati P, Moran MA . (2010). Genes for transport and metabolism of spermidine in Ruegeria pomeroyi DSS-3 and other marine bacteria. Aquat Microb Ecol 58: 311–321.
Moustafa A, Evans AN, Kulis DM, Hackett JD, Erdner DL, Anderson DM et al. (2010). Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS ONE 5: e9688.
Natrah FMI, Kenmegne MM, Wiyoto W, Sorgeloos P, Bossier P, Defoirdt T . (2011). Effects of micro-algae commonly used in aquaculture on acyl-homoserine lactone quorum sensing. Aquaculture 317: 53–57.
Needham DM, Fuhrman JA . (2016). Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol 1: 16005.
Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE et al. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME J 4: 784–798.
Nishibori N, Nishio S . (1997). Occurrence of polyamines in the bloom forming toxic dinoflagellate Alexandrium tamarense. Fish Sci 63: 319–320.
Nishibori N, Yuasa A, Sakai M, Fujihara S, Nishio S . (2001). Free polyamine concentrations in coastal seawater during phytoplankton bloom. Fish Sci 67: 79–83.
Parker MS, Armbrust EV . (2005). Synergistic effects of light, temperature, and nitrogen source on transcription of genes for carbon and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae). J Phycol 41: 1142–1153.
Poretsky RS, Sun S, Mou X, Moran MA . (2010). Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol 12: 616–627.
Reisch CR, Moran MA, Whitman WB . (2008). Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J Bacteriol 190: 8018–8024.
Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB . (2011). Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473: 208–211.
Rivers AR, Smith CB, Moran MA . (2014). An updated genome annotation for the model marine bacterium Ruegeria pomeroyi DSS-3. Stand Genomic Sci 9: 11.
Ryan JP, McManus MA, Kudela RM, Lara Artigas M, Bellingham JG, Chavez FP et al. (2014). Boundary influences on HAB phytoplankton ecology in a stratification-enhanced upwelling shadow. Deep Sea Res Part II Top Stud Oceanogr 101: 63–79.
Schäfer H, Abbas B, Witte H, Muyzer G . (2002). Genetic diversity of ‘satellite’ bacteria present in cultures of marine diatoms. FEMS Microbiol Ecol 42: 25–35.
Schulz A, Stöveken N, Binzen IM, Hoffmann T, Heider J, Bremer E . (2017). Feeding on compatible solutes: a substrate-induced pathway for uptake and catabolism of ectoines and its genetic control by EnuR. Environ Microbiol 19: 926–946.
Seeyave S, Probyn TA, Pitcher GC, Lucas MI, Purdie DA . (2009). Nitrogen nutrition in assemblages dominated by Pseudo-nitzschia spp., Alexandrium catenella and Dinophysis acuminata off the west coast of South Africa. Mar Ecol Prog Ser 379: 91–107.
Segev E, Wyche TP, Kim KH, Petersen J, Ellebrandt C, Vlamakis H et al. (2016). Dynamic metabolic exchange governs a marine algal-bacterial interaction. eLife 5: e17473.
Seyedsayamdost MR, Case RJ, Kolter R, Clardy J . (2011). The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem 3: 331–335.
Sison-Mangus MP, Jiang S, Tran KN, Kudela RM . (2014). Host-specific adaptation governs the interaction of the marine diatom, Pseudo-nitzschia and their microbiota. ISME J 8: 63–76.
Smriga S, Fernandez VI, Mitchell JG, Stocker R . (2016). Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc Natl Acad Sci 113: 1576–1581.
Stewart FJ, Ottesen EA, DeLong EF . (2010). Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J 4: 896–907.
Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF . (2008). Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci 105: 4209–4214.
Sunda WG, Hardison DR . (2008). Contrasting seasonal patterns in dimethylsulfide, dimethylsulfoniopropionate, and chlorophyll a in a shallow North Carolina estuary and the Sargasso Sea. Aquat Microb Ecol 53: 281–294.
Taylor FJR, Hoppenrath M, Saldarriaga JF . (2008). Dinoflagellate diversity and distribution. Biodivers Conserv 17: 407–418.
Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611.
Todd JD, Curson ARJ, Sullivan MJ, Kirkwood M, Johnston AWB . (2012a). The Ruegeria pomeroyi acuI gene has a role in DMSP catabolism and resembles yhdH of E. coli and other bacteria in conferring resistance to acrylate. PLoS ONE 7: e35947.
Todd JD, Kirkwood M, Newton-Payne S, Johnston AWB . (2012b). DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J 6: 223–226.
Varaljay VA, Robidart J, Preston CM, Gifford SM, Durham BP, Burns AS et al. (2015). Single-taxon field measurements of bacterial gene regulation controlling DMSP fate. ISME J 9: 1677–1686.
Wehrens R, Buydens LM . (2007). Self-and super-organizing maps in R: the Kohonen package. J Stat Softw 21: 1–19.
Wiegmann K, Hensler M, Wöhlbrand L, Ulbrich M, Schomburg D, Rabus R . (2014). Carbohydrate catabolism in Phaeobacter inhibens DSM 17395, a member of the marine Roseobacter clade. Appl Environ Microbiol 80: 4725–4737.
Williams P . (2007). Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153: 3923–3938.
Wright RT, Shah NM . (1977). The trophic role of glycolic acid in coastal seawater. II. Seasonal changes in concentration and heterotrophic use in Ipswich Bay, Massachusetts, USA. Mar Biol 43: 257–263.
Zan J, Heindl JE, Liu Y, Fuqua C, Hill RT . (2013). The CckA-ChpT-CtrA phosphorelay system is regulated by quorum sensing and controls flagellar motility in the marine sponge symbiont Ruegeria sp. KLH11. PLOS One 8: e66346.
Acknowledgements
We appreciate advice and assistance from B Nowinski, S Sharma, C Smith, B Durham, and A Vorobev. This work was funded by NSF grant OCE-1342694 and Gordon and Betty Moore Foundation grant #5503.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Landa, M., Burns, A., Roth, S. et al. Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J 11, 2677–2690 (2017). https://doi.org/10.1038/ismej.2017.117
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.117
This article is cited by
-
Regulation of DMSP organosulfur cycling in ubiquitous Roseobacter marine bacteria
The EMBO Journal (2026)
-
A review of quorum-sensing and its role in mediating interkingdom interactions in the ocean
Communications Biology (2025)
-
A high-resolution diel survey of surface ocean metagenomes, metatranscriptomes, and transfer RNA transcripts
Scientific Data (2025)
-
Digital Microbe: a genome-informed data integration framework for team science on emerging model organisms
Scientific Data (2024)
-
Bacterial transcriptional response to labile exometabolites from photosynthetic picoeukaryote Micromonas commoda
ISME Communications (2023)