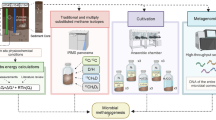
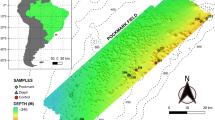
Abstract
Despite accounting for the majority of sedimentary methane, the physiology and relative abundance of subsurface methanogens remain poorly understood. We combined intact polar lipid and metagenome techniques to better constrain the presence and functions of methanogens within the highly reducing, organic-rich sediments of Antarctica’s Adélie Basin. The assembly of metagenomic sequence data identified phylogenic and functional marker genes of methanogens and generated the first Methanosaeta sp. genome from a deep subsurface sedimentary environment. Based on structural and isotopic measurements, glycerol dialkyl glycerol tetraethers with diglycosyl phosphatidylglycerol head groups were classified as biomarkers for active methanogens. The stable carbon isotope (δ13C) values of these biomarkers and the Methanosaeta partial genome suggest that these organisms are acetoclastic methanogens and represent a relatively small (0.2%) but active population. Metagenomic and lipid analyses suggest that Thaumarchaeota and heterotrophic bacteria co-exist with Methanosaeta and together contribute to increasing concentrations and δ13C values of dissolved inorganic carbon with depth. This study presents the first functional insights of deep subsurface Methanosaeta organisms and highlights their role in methane production and overall carbon cycling within sedimentary environments.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ et al.(2014). Binning metagenomic contigs by coverage and composition. Nat Methods 11: 1144–1146.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards R et al.. (2008). The RAST server: Rapid Annotations using Subsystems Technology. BMC Genomics 9: 1–15.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al.(2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477.
Barber RD, Zhang L, Harnack M, Olson MV, Kaul R, Ingram-Smith C et al.(2011). Complete genome sequence of Methanosaeta concilii, a specialist in aceticlastic methanogenesis. J Bacteriol 193: 3668–3669.
Becker KW, Lipp JS, Zhu C, Liu X-L, Hinrichs K-U . (2013). An improved method for analysis of archaeal and bacterial ether core lipids. Org Geochem 61: 34–44.
Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R et al.(2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. PNAS 103: 3846–3851.
Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH . (2008). Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. PNAS 105: 10583–10588.
Blair NE, Carter WD . (1992). The carbon isotope biogeochemistry of acetate from a methanogenic marine sediment. Geochim Cosmochim Acta 56: 1247–1258.
Blumenberg M, Seifert R, Reitner J, Pape T, Michaelis W . (2004). Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. PNAS 101: 11111–11116.
Bray NL, Pimentel H, Melsted P, Pachter L . (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotech 34: 525–527.
Burdige DJ . (2006) Geochemistry of Marine Sediments. Princeton University Press: Princeton, NJ, USA.
Bushnell B . (2015) BBMap. Available at: http://sourceforge.net/projects/bbmap/.
Carr SA, Orcutt BN, Mandernack KW, Spear JR . (2015). Abundant Atribacteria in deep marine sediment from the Adélie Basin, Antarctica. Front Microbiol 6: 1–12.
Carr SA, Mills CT, Mandernack KW . (2016). The use of amino acid indices for assessing organic matter quality and microbial abundance in deep-sea Antarctic sediments of IODP Expedition 318. Mar Chem 186: 72–82.
Carr SA, Vogel SW, Dunbar RB, Brandes J, Spear JR, Levy R et al.(2013). Bacterial abundance and composition in marine sediments beneath the Ross Ice Shelf, Antarctica. Geobiology 11: 377–395.
Colwell FS, Boyd S, Delwiche ME, Reed DW, Phelps TJ, Newby DT . (2008). Estimates of biogenic methane production rates in deep marine sediments at Hydrate Ridge, Cascadia Margin. Appl Environ Microb 74: 3444–3452.
Dodsworth JA, Blainey PC, Murugapiran SK, Swingley WD, Ross CA, Tringe SG et al.(2013). Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nat Commun 4: 1810–1854.
Elling FJ, Könneke M, Mußmann M, Greve A, Hinrichs K-U . (2015). Influence of temperature, pH, and salinity on membrane lipid composition and TEX86 of marine planktonic thaumarchaeal isolates. Geochim Cosmochim Acta 171: 238–255.
Elling FJ, Könneke M, Nicol GW, Stieglmeier M, Bayer B, Spieck E et al.(2017). Chemotaxonomic characterisation of the thaumarchaeal lipidome. Environ Microb 171: 238–255.
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML et al.(2015). Anvi’o: an advanced analysis and visualization platform for ‘omics data’. PeerJ 3: e1319.
Expedition 318 Scientists. (2011a) Methods. In: Escutia C, Brinkhuis H, Klaus A the Expedition 318 Scientists(eds.)Proc. IODP, 318. Integrated Ocean Drilling Program Management International, Inc: Tokyo, Japan.
Expedition 318 Scientists. (2011b) Site U1357. In: Escutia C, Brinkhuis H, Klaus A the Expedition 318 Scientists(eds.)Proc. IODP, 318. Integrated Ocean Drilling Program Management International, Inc.: Tokyo, Japan.
Ferdelman TG, Lee C, Pantoja S, Harder J, Bebout BM, Fossing H . (1997). Sulfate reduction and methanogenesis in a Thioploca-dominated sediment off the coast of Chile. Geochim Cosmochim Acta 61: 3065–3079.
Ferry JG, Kastead KA . (2007) Methanogenesis. In: Cavicchioli R(ed.)Archaea: Molecular and Cellular Biology. ASM Press: Washington, DC, USA, pp288–314.
Ferry JG, Lesser DJ . (2008). Methanogenesis in marine sediments. Ann NY Acad Sci 1125: 147–157.
Fitzsimons MF, Jemmett AW, Wolff GA . (1997). A preliminary study of the geochemistry of methylamines in a salt marsh. Org Geochem 27: 15–24.
Gies EA, Konwar KM, Beatty JT, Hallam SJ . (2014). Illuminating microbial dark matter in Meromictic Sakinaw Lake. Appl Environ Microb 80: 6807–6818.
Gruber N, Keeling CD, Bacastow RB, Guenther PR, Lueker TJ, Wahlen M et al.(1999). Spatiotemporal patterns of carbon-13 in the global surface oceans and the oceanic suess effect. Global Biogeochem Cy 13: 307–335.
Heuer VB, Krüger M, Evert M, Hinrichs K-U . (2010). Experimental studies on the stable carbon isotope biogeochemistry of acetate in lake sediments. Org Geochem 41: 22–30.
Heuer VB, Pohlman JW, Torres ME, Elvert M . (2009). The stable carbon isotope biogeochemistry of acetate and other dissolved carbon species in deep subseafloor sediments at the northern Cascadia Margin. Geochim Cosmochim Acta 73: 3323–3336.
Hopmans EC, Weijers JWH, Schefuß E, Herfort L, Sinninghe Damsté JS, Schouten S . (2004). A novel proxy for terrestrial organic matter in sediments based on branched and isoprenoid tetraether lipids. Earth Planet Sci Lett 224: 107–116.
Huang Y, Gilna P, Li W . (2009). Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25: 1338–1340.
Huguet C, Hopmans EC, Febo-Ayala W, Thompson DH, Sinninghe Damsté JS, Schouten S . (2006), An improved method to determine the absolute abundance of glycerol dibiphytanyl glycerol tetraether lipids. Org Geochem 37:1036–1041.
Hurley SJ, Elling FJ, Könneke M, Buchwald C, Wankel SD, Santoro AE et al.(2016). Influence of ammonia oxidation rate on thaumarchaeal lipid composition and the TEX86 temperature proxy. PNAS 113: 7762–7767.
Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A et al.(2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. PNAS 103: 2815–2820.
Jahn U, Summons R, Sturt H, Grosjean E, Huber H . (2004). Composition of the lipids of Nanoarchaeum equitans and their origin from its host Ignicoccus sp. strain KIN4/I. Arch Microbiol 182: 404–413.
Katayama T, Yoshioka H, Takahashi HA, Amo M, Fujii T, Sakata S . (2016). Changes in microbial communities associated with gas hydrates in subseafloor sediments from the Nankai Trough. FEMS Microbiol Ecol 92:1–10.
Koga Y, Nakano M . (2008). A dendrogram of archaea based on lipid component parts composition and its relationship to rRNA phylogeny. Syst Appl Microbiol 31: 169–182.
Könneke M, Lipp JS, Hinrichs K-U . (2012). Carbon isotope fractionation by the marine ammonia-oxidizing archaeon Nitrosopumilus maritimus. Org Geochem 48: 21–24.
Lang K, Klingl A, Poehlein A, Daniel R, Brune A . (2015). New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of ‘Candidatus Methanoplasma termitum’. Appl Environ Microb 81: 1338–1352.
Lengger SK, Hopmans EC, Reichart G-J, Nierop KGJ, Sinninghe Damsté JS, Schouten S . (2012). Intact polar and core glycerol dibiphytanyl glycerol tetraether lipids in the Arabian Sea oxygen minimum zone. Part II: Selective preservation and degradation in sediments and consequences for the TEX86. Geochim Cosmochim Acta 98: 244–258.
Leventer A, Domack E, Dunbar R, Pike J, Stickley C, Maddison E et al.. (2006). Marine sediment record from the East Antarctic margin reveals dynamics of ice sheet recession. GSA Today 16: 4–10.
Li D, Liu C-M, Luo R, Sadakane K, Lam T-W . (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31: 1674–1676.
Liang B, Wang LY, Mbadinga SM, Liu JF, Yang SZ, Gu JD et al.(2015). Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Express 5:1–13.
Lipp JS, Hinrichs K-U . (2009). Structural diversity and fate of intact polar lipids in marine sediments. Geochim Cosmochim Acta 73: 6816–6833.
Lipp JS, Morono Y, Inagaki F, Hinrichs K-U . (2008). Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454: 991–994.
Liu X-L, Lipp JS, Schröder JM, Summons RE, Hinrichs K-U . (2012). Isoprenoid glycerol dialkanol diethers: a series of novel archaeal lipids in marine sediments. Org Geochem 43: 50–55.
Londry KL, Dawson KG, Grover HD, Summons RE, Bradley AS . (2008). Stable carbon isotope fractionation between substrates and products of Methanosarcina barkeri. Org Geochem 39: 608–621.
Lovell CR, Przybyla A, Ljungdahl LG . (1990). Primary structure of the thermostable formyltetrahydrofolate synthetase from Clostridium thermoaceticum. Biochemistry 29: 5687–5694.
Ma K, Liu X, Dong X . (2006). Methanosaeta harundinacea sp. nov., a novel acetate-scavenging methanogen isolated from a UASB reactor. Int J Syst Evol Microbiol 56: 127–131.
Markowitz VM, Chen I-MA, Palaniapan K, Chu k, Szeto E, Pillay M et al.(2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleci Acids Res 43: D560–D567.
Miller CS, Baker BJ, Thomas BC, Singer SW, Banfield JF . (2011). EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol 12: 1–14.
Mills CT, Slater GF, Dias RF, Carr SA, Reddy CM, Schmidt R et al.(2013). The relative contribution of methanotrophs to microbial communities and carbon cycling in soil overlying a coal-bed methane seep. FEMS Microbiol Ecol 84: 474–494.
Mook WG, Bommerson JC, Staverman WH . (1974). Carbon isotope fractionation between dissolved bicarbonate and gaseous carbon dioxide. Earth Planet Sci Lett 22: 169–176.
Mori K, Iino T, Suzuki KI, Yamaguchi K, Kamagata Y . (2012). Aceticlastic and NaCl-requiring methanogen ‘Methanosaeta pelagica’ sp. Nov., isolated from marine tidal flat sediment. Appl Environ Microb 78: 3416–3423.
Nobu MK, Narihiro T, Rinke C, Kamagata Y, Tringe SG, Woyke T et al.(2015). Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 9: 1710–1722.
Pancost RD, Sinninghe Damsté JS . (2003). Carbon isotopic compositions of prokaryotic lipids as tracers of carbon cycling in diverse settings. Chem Geol 195: 29–58.
Parkes RJ, Webster G, Cragg BA, Weightman AJ, Newberry CJ, Ferdelman TG et al.(2005). Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 436: 390–394.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW . (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055.
Patel GB . (1984). Characterization and nutritional properties of Methanothrix concilii sp. nov., a mesophilic, aceticlastic methanogen. Can J Microbiol 30: 1383–1396.
Paull CK, Lorenson TD, Borowski WS, Ussler W, Olsen K, Rodriguez NM . (2000) Isotopic composition of CH4 and CO2 species, and sedimentary organic matter within samples from the Blake Ridge: gas source implications. In: Paull CK, Matsumoto R, Wallace PJ, Dillon WP(eds.)Proceedings of the Ocean Drilling Program, Scientific Results. Ocean Drilling Program: College Station, TX, USA, 164,pp 67–78.
Pearson A, Hurley SJ, Walter SRS, Kusch S, Lichtin S, Zhang YG . (2016). Stable carbon isotope ratios of intact GDGTs indicate heterogeneous sources to marine sediments. Geochim Cosmochim Acta 181: 18–35.
Pitcher A, Hopmans EC, Mosier AC, Park S-J, Rhee S-K, Francis CA et al.. (2011). Core and intact polar glycerol dibiphytanyl glycerol tetraether lipids of ammonia-oxidizing archaea enriched from marine and estuarine sediments. Appl Environ Microb 77: 3468–3477.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al.(2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596.
Qin W, Amin SA, Martens-habbena W, Walker CB, Urakawa H, Devol AH . (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. PNAS 111: 12504–12509.
Reeburgh WS . (2007) Oceanic methane biogeochemistry. Chem Rev 107: 486–513.
Rodriguez-R LM, Konstantinidis KT . (2014). Nonpareil: a redundancy-based approach to assess the level of coverage in metagenomic datasets. Bioinformatics 30: 629–635.
Rotaru AE, Shrestha D, Embree M, Zengler K, Wardman C . (2014). A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci 7: 408–415.
Schouten S, Hopmans EC, Baas M, Boumann H, Standfest S, Könneke M et al.(2008). Intact membrane lipids of ‘Candidatus Nitrosopumilus maritimus,’ a cultivated representative of the cosmopolitan mesophilic group I Crenarchaeota. Appl Environ Microb 74: 2433–2440.
Schouten S, Middelburg JJ, Hopmans EC, Sinninghe Damsté JS . (2010). Fossilization and degradation of intact polar lipids in deep subsurface sediments: a theoretical approach. Geochim Cosmochim Acta 74: 3806–3814.
Schubotz F, Wakeham SG, Lipp JS, Fredricks HF, Hinrichs K . (2009). Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environ Microbiol 11: 2720–2734.
Schubotz F, Lipp JS, Elvert M, Hinrichs K-U . (2011). Stable carbon isotopic compositions of intact polar lipids reveal complex carbon flow patterns among hydrocarbon degrading microbial communities at the Chapopote asphalt volcano. Geochim Cosmochim Acta 75: 4399–4415.
Schubotz F, Meyer-Dombard DR, Bradley AS, Fredricks HF, Hinrichs K-U, Shock EL et al.(2013). Spatial and temporal variability of biomarkers and microbial diversity reveal metabolic and community flexibility in Streamer Biofilm Communities in the Lower Geyser Basin, Yellowstone National Park. Geobiology 11: 549–569.
Smith KS, Ingram-smith C . (2007). Methanosaeta, the forgotten methanogen? Trends Microbiol 15: 150–155.
Sowers KR, Baron SF, Ferry JG . (1984). Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl Environ Microb 47: 971–978.
Sowers KR, Ferry JG . (2003) Methanogenesis in the Marine Environment. Encyclopedia of Environmental Microbiology. John Wiley & Sons, Inc.: New York, NY, USA..
Takano Y, Chikaraishi Y, Ogawa NO, Nomaki H, Morono Y, Inagaki F et al.. (2010). Sedimentary membrane lipids recycled by deep-sea benthic archaea. Nat Geosci 3: 1–4.
Teece MA, Fogel ML, Dollhopf ME, Nealson KH . (1999). Isotopic fractionation associated with biosynthesis of fatty acids by a marine bacterium under oxic and anoxic conditions. Org Geochem 30: 1571–1579.
Torres ME, Kastner M . (2009) Data report: clues about carbon cycling in methane-bearing sediments using stable isotopes of the dissolved inorganic carbon, IODP Expedition 311. In: Riedel M, Collett TS, Malone MJ the Expedition 311 Scientists(eds.)Proc. IODP 311. Integrated Ocean Drilling Program Management International, Inc.: Washington, DC, USA.
Valentine DL . (2002). Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review. Van Leeuw J Microb 81: 271–282.
Valentine DL . (2011). Emerging topics in marine methane biogeochemistry. Annu Rev Mar Sci 3: 147–171.
Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ et al.. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. PNAS 107: 8818–8823.
Wang Y, Coleman-Derr D, Chen G, Gu YQ . (2015). OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 43: W78–W84.
Wehrmann LM, Risgarrd-Petersen N, Schrum HN, Walsh EA, Huh Y, Ikehara M et al.(2011). Coupled organic and inorganic carbon cycling in the deep subseafloor sediment of the northeastern Bering Sea Slope (IODP Exp. 323). Chem Geol 284: 251–261.
Whiticar M, Faber E, Schoel ML . (1986). Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation-isotope evidence. Geochim Cosmochim Acta 50: 693–709.
Wörmer L, Lipp JS, Schröder JM, Hinrichs K-U . (2013). Application of two new LC-ESI-MS methods for improved detection of intact polar lipids (IPLs) in environmental samples. Org Geochem 59: 10–21.
Xie S, Lipp JS, Wegener G, Ferdelman TG, Hinrichs K-U . (2013). Turnover of microbial lipids in the deep biosphere and growth of benthic archaeal populations. PNAS 110: 6010–6014.
Xie S, Pancost RD, Chen L, Evershed RP, Yang H, Zhang K et al.(2012). Microbial lipid records of highly alkaline deposits and enhanced aridity associated with significant uplift of the Tibetan Plateau in the Late Miocene. Geology 40: 291–294.
Yoshinaga MY, Kellermann MY, Rossel PE, Schubotz F, Lipp JS, Hinrichs K-U . (2011). Systematic fragmentation patterns of archaeal intact polar lipids by high-performance liquid chromatography/electrospray ionization ion-trap mass spectrometry. Rapid Commun Mass Sp 25: 3563–3574.
Yoshinaga MY, Lazar CS, Elvert M, Lin Y, Zhu C, Heuer VB et al.(2015). Possible roles of uncultured archaea in carbon cycling in methane-seep sediments. Geochim Cosmochim Acta 164: 35–52.
Zhang YG, Zhang CL, Liu X-L, Li L, Hinrichs K-U, Noakes JE . (2011). Methane Index: a tetraether archaeal lipid biomarker indicator for detecting the instability of marine gas hydrates. Earth Planet Sci Lett 307: 525–534.
Zhou J, Bruns MA, Tiedje JM . (1996). DNA recovery from soils of diverse composition. Appl Environ Microb 62: 316–322.
Zhu C, Lipp JS, Wörmer L, Becker KW, Schröder J, Hinrichs K-U . (2013). Comprehensive glycerol ether lipid fingerprints through a novel reversed phase liquid chromatography–mass spectrometry protocol. Org Geochem 65: 53–62.
Zink K-G, Wilkes H, Disko U, Elvert M, Horsfield B . (2003). Intact phospholipids—microbial ‘life markers’ in marine deep subsurface sediments. Org Geochem 34: 755–769.
Acknowledgements
We thank the shipboard party of IODP Expedition 318 for supporting this work, especially co-chiefs Escutia and Brinkhuis, Staff Scientist Klaus and geochemistry team: James Bendle, Tina van de Flierdt, and Francisco Jimenez-Espejo. We thank Charles Pepe-Ranney and Benjamin Tully for bioinformatics assistance, David Mucciarone for isotope geochemistry assistance, Kai-Uwe Hinrichs and John Spear for the use of their facilities, Julius Lipp for provision of IPL standards and Beth Orcutt for manuscript comments. The Deep Carbon Observatory’s Census of Deep Life project funded by the Alfred P Sloan Foundation provided sequencing services. Carr was funded by the US National Science Foundation (NSF OCE-0939564) and the Center for Dark Energy Biosphere Investigations (funded by NSF OCE-1521614, publication no. 390). FS and RES acknowledge funding from NASA Astrobiology grant (NNA13AA90A) and the Central Research Development Fund, University of Bremen. Any use of trade, firm or product names is for descriptive purposes only and does not imply endorsement by the US Government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Carr, S., Schubotz, F., Dunbar, R. et al. Acetoclastic Methanosaeta are dominant methanogens in organic-rich Antarctic marine sediments. ISME J 12, 330–342 (2018). https://doi.org/10.1038/ismej.2017.150
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.150
This article is cited by
-
Distribution of Microbial Community in Coal Reservoir with Ability to Generate Methane by Anaerobically Degrading Coal
Water, Air, & Soil Pollution (2025)
-
Metagenomic insights into the impacts of phytochemicals on bacterial and archaeal community structure and biogas production patterns during anaerobic digestion of avocado oil processing waste feedstocks
Biomass Conversion and Biorefinery (2024)
-
Microbial community evolution in a lab-scale reactor operated to obtain biomass for biochemical methane potential assays
Applied Microbiology and Biotechnology (2024)
-
Microbial life in 25-m-deep boreholes in ancient permafrost illuminated by metagenomics
Environmental Microbiome (2023)
-
Vertically stratified methane, nitrogen and sulphur cycling and coupling mechanisms in mangrove sediment microbiomes
Microbiome (2023)