Abstract
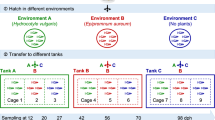
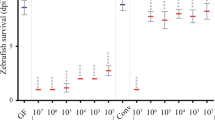
All animals live in intimate association with communities of microbes, collectively referred to as their microbiota. Certain host traits can influence which microbial taxa comprise the microbiota. One potentially important trait in vertebrate animals is the adaptive immune system, which has been hypothesized to act as an ecological filter, promoting the presence of some microbial taxa over others. Here we surveyed the intestinal microbiota of 68 wild-type zebrafish, with functional adaptive immunity, and 61 rag1− zebrafish, lacking functional B- and T-cell receptors, to test the role of adaptive immunity as an ecological filter on the intestinal microbiota. In addition, we tested the robustness of adaptive immunity’s filtering effects to host–host interaction by comparing the microbiota of fish populations segregated by genotype to those containing both genotypes. The presence of adaptive immunity individualized the gut microbiota and decreased the contributions of neutral processes to gut microbiota assembly. Although mixing genotypes led to increased phylogenetic diversity in each, there was no significant effect of adaptive immunity on gut microbiota composition in either housing condition. Interestingly, the most robust effect on microbiota composition was co-housing within a tank. In all, these results suggest that adaptive immunity has a role as an ecological filter of the zebrafish gut microbiota, but it can be overwhelmed by other factors, including transmission of microbes among hosts.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J et al. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 107: 18933–18938.
Brugman S, Schneeberger K, Witte M, Klein MR, van den Bogert B, Boekhorst J et al. (2014). T lymphocytes control microbial composition by regulating the abundance of Vibrio in the zebrafish gut. Gut Microbes 5: 737–747.
Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD et al. (2015). Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 43: 541–553.
Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K et al. (2016). Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J 10: 655–664.
Daniels JA, Lederman HM, Maitra A, Montgomery EA . (2007). Gastrointestinal tract pathology in patients with common variable immunodeficiency (CVID). Am J Surg Pathol 31: 1800–1812.
Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, Gilad Y . (2015). Genome-wide association studies of the human gut microbiota. PLoS ONE 10: e0140301.
Dimitriu PA, Boyce G, Samarakoon A, Hartmann M, Johnson P, Mohn WW . (2013). Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environ Microbiol Rep 5: 200–210.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Faith DP . (1992). Conservation evaluation and phylogenetic diversity. Biol Conserv 61: 1–10.
Flajnik MF, Kasahara M . (2010). Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11: 47–59.
Fox J, Weisberg S . (2011) An {R} Companion to Applied Regression. 2nd edition.Sage: Thousand Oaks, CA, USA.
Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S et al. (2015). BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity 43: 527–540.
George LO, Bazzaz FA . (1999). The fern understory as an ecological filter: emergence and establishment of canopy-tree seedlings. Ecology 80: 833–845.
Goodrich JK, Davenport ER, Waters JL, Clark AG, Ley RE . (2016). Cross-species comparisons of host genetic associations with the microbiome. Science 352: 532–535.
Gori A, Tincati C, Rizzardini G, Torti C, Quirino T, Haarman M et al. (2008). Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J Clin Microbiol 46: 757–758.
Hannon Lab. (2010).FASTX Toolkit. Available from http://hannonlab.cshl.edu/fastx_toolkit/index.html.
Hohn C, Petrie-Hanson L . (2012). Rag1−/− mutant zebrafish demonstrate specific protection following bacterial re-exposure. PLoS ONE 7: e44451.
Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC . (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA 105: 15064–15069.
Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y et al. (2014). Foxp3+ T cells regulate immunoglobulin A selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41: 152–165.
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464.
Kostic AD, Howitt MR, Garrett WS . (2013). Exploring host-microbiota interactions in animal models and humans. Genes Dev 27: 701–718.
Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O’Connell RM et al. (2015a). MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 17: 153–163.
Kubinak JL, Stephens WZ, Soto R, Petersen C, Chiaro T, Gogokhia L et al. (2015b). MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun 6: 8642.
Lam S, Chua HL, Gong Z, Lam TJ, Sin YM . (2004). Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev Comp Immunol 28: 9–28.
Lau MK . (2013).DTK: Dunnett-Tukey-Kramer Pairwise Multiple Comparison Test Adjusted for Unequal Variances and Unequal Sample Sizes. Available from https://cran.r-project.org/package=DTK.
Love MI, Huber W, Anders S . (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Magoc T, Salzberg SL . (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963.
McMurdie PJ, Holmes S . (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8: e61217.
McMurdie PJ, Holmes S . (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10: e1003531.
Mouquet N, Loreau M . (2002). Coexistence in metacommunities: the regional similarity hypothesis. Am Nat 159: 420–426.
Oh J, Freeman AF, Park M, Sokolic R, Candotti F, Holland SM et al. (2013). The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res 23: 2103–2114.
Oksanen J, Blanchet FGG, Kindt R, Legendre P, Minchin PRR, O’Hara RBB et al. (2016).vegan: Community Ecology Package. Available from https://cran.r-project.org/package=vegan.
Org E, Parks BW, Joo JWJ, Emert B, Schwartzman W, Kang EY et al. (2015). Genetic and environmental control of host-gut microbiota interactions. Genome Res 25: 1558–1569.
Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158: 1000–1010.
R Core Team. (2015). R: A Language and Environment for Statistical Computing. Available from https://www.r-project.org/.
Rauta PR, Nayak B, Das S . (2012). Immune system and immune responses in fish and their role in comparative immunity study: a model for higher organisms. Immunol Lett 148: 23–33.
Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K et al. (2011). Evidence for a core gut microbiota in the zebrafish. ISME J 5: 1595–1608.
Shen W, Li W, Hixon JA, Bouladoux N, Belkaid Y, Dzutzev A et al. (2014). Adaptive immunity to murine skin commensals. Proc Natl Acad Sci USA 111: E2977–E2986.
Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP . (2006). Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol 8: 732–740.
Sørensen T . (1948). A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analysis of the vegetation on Danish commons. Biol Skr 5: 1–34.
Spor A, Koren O, Ley R . (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9: 279–290.
Stephens WZ, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K et al. (2016). The composition of the zebrafish intestinal microbial community varies across development. ISME J 10: 644–654.
Thoene-Reineke C, Fischer A, Friese C, Briesemeister D, Göbel UB, Kammertoens T et al. (2014). Composition of intestinal microbiota in immune-deficient mice kept in three different housing conditions. PLoS ONE 9: e113406.
Traver D, Herbomel P, Patton EE, Murphey RD, Yoder JA, Litman GW et al. (2003). The zebrafish as a model organism to study development of the immune system. Adv Immunol 81: 254–330.
Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI . (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3: 213–223.
Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR . (2009). High-throughput sequencing of the zebrafish antibody repertoire. Science 324: 807–810.
Wickham H . (2009) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag: New York, NY, USA.
Wienholds E . (2002). Target-selected inactivation of the zebrafish rag1 gene. Science 297: 99–102.
Wong S, Stephens WZ, Burns AR, Stagaman K, David LA, Bohannan BJM et al. (2015). Ontogenetic differences in dietary fat influence microbiota assembly in the zebrafish gut. MBio 6: e00687–15.
Zhang H, Sparks JB, Karyala SV, Settlage R, Luo XM . (2015). Host adaptive immunity alters gut microbiota. ISME J 9: 770–781.
Acknowledgements
Research reported in this publication was supported by the National Institute of General Medical Sciences of the NIH under award numbers R01GM095385 and P50GM098911. Grant P01HD22486 provided support for the Oregon Zebrafish Facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The ACISS computational resources were funded by a Major Research Instrumentation grant (Grant No. OCI-0960354) from the National Science Foundation, Office of Cyber Infrastructure. We also thank Rose Sockol for crossing and maintaining zebrafish in the Oregon Zebrafish Facility.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Stagaman, K., Burns, A., Guillemin, K. et al. The role of adaptive immunity as an ecological filter on the gut microbiota in zebrafish. ISME J 11, 1630–1639 (2017). https://doi.org/10.1038/ismej.2017.28
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.28
This article is cited by
-
Bridging Ecology and Microbiomes: Applying Ecological Theories in Host-associated Microbial Ecosystems
Current Clinical Microbiology Reports (2025)
-
Unravelling the temporal and spatial variation of fungal phylotypes from embryo to adult stages in Atlantic salmon
Scientific Reports (2024)
-
Collembolans maintain a core microbiome responding to diverse soil ecosystems
Soil Ecology Letters (2024)
-
The interactions between the host immunity and intestinal microorganisms in fish
Applied Microbiology and Biotechnology (2024)
-
Robust host source tracking building on the divergent and non-stochastic assembly of gut microbiomes in wild and farmed large yellow croaker
Microbiome (2022)