Abstract
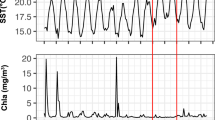
Numerous ecological processes, such as bacteriophage infection and phytoplankton–bacterial interactions, often occur via strain-specific mechanisms. Therefore, studying the causes of microbial dynamics should benefit from highly resolving taxonomic characterizations. We sampled daily to weekly over 5 months following a phytoplankton bloom off Southern California and examined the extent of microdiversity, that is, significant variation within 99% sequence similarity clusters, operational taxonomic units (OTUs), of bacteria, archaea, phytoplankton chloroplasts (all via 16S or intergenic spacer (ITS) sequences) and T4-like-myoviruses (via g23 major capsid protein gene sequence). The extent of microdiversity varied between genes (ITS most, g23 least) and only temporally common taxa were highly microdiverse. Overall, 60% of taxa exhibited microdiversity; 59% of these had subtypes that changed significantly as a proportion of the parent taxon, indicating ecologically distinct taxa. Pairwise correlations between prokaryotes and myoviruses or phytoplankton (for example, highly microdiverse Chrysochromulina sp.) improved when using single-base variants. Correlations between myoviruses and SAR11 increased in number (172 vs 9, Spearman>0.65) and became stronger (0.61 vs 0.58, t-test: P<0.001) when using SAR11 ITS single-base variants vs OTUs. Whole-community correlation between SAR11 and myoviruses was much improved when using ITS single-base variants vs OTUs, with Mantel rho=0.49 vs 0.27; these results are consistent with strain-specific interactions. Mantel correlations suggested >1 μm (attached/large) prokaryotes are a major myovirus source. Consideration of microdiversity improved observation of apparent host and virus networks, and provided insights into the ecological and evolutionary factors influencing the success of lineages, with important implications to ecosystem resilience and microbial function.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL et al. (2004). Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430: 551–554.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101.
Amin SA, Parker MS, Armbrust EV . (2012). Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76: 667–684.
Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D . (2011). Genomic island variability facilitates Prochlorococcus–virus coexistence. Nature 474: 604–608.
Bettarel Y, Motegi C, Weinbauer MG, Mari X . (2015). Colonization and release processes of viruses and prokaryotes on artificial marine macroaggregates. FEMS Microbiol Lett 363: 1–8.
Bolger AM, Lohse M, Usadel B . (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120.
Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D et al. (2002). Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA 99: 14250–14255.
Brown CT . (2015). Strain recovery from metagenomes. Nat Biotechnol 33: 1041–1043.
Brown MV, Fuhrman JA . (2005). Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat Microb Ecol 41: 15–23.
Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T et al. (2012). Global biogeography of SAR11 marine bacteria. Mol Syst Biol 8: 595.
Brown MV, Schwalbach MS, Hewson I, Fuhrman JA . (2005). Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ Microbiol 7: 1466–1479.
Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP . (2016). DADA2: high resolution sample inference from amplicon data. Nat Methods 13: 581–583.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Chow C-ET, Fuhrman JA . (2012). Seasonality and monthly dynamics of marine myovirus communities. Environ Microbiol 14: 2171–2183.
Chow C-ET, Kim DY, Sachdeva R, Caron DA, Fuhrman JA . (2014). Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J 8: 816–829.
Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P et al. (2014). Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513: 242–245.
Ducklow HW, Steinberg DK, Buesseler KO . (2001). Upper ocean carbon export and the biological pump. Oceanography 14: 50–58.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Edgar RC . (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998.
Eren AM, Borisy GG, Huse SM, Mark Welch JL . (2014). Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA 111: E2875–E2884.
Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG et al. (2013). Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4: 1111–1119.
Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML . (2014). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9: 968–979.
Filée J, Tetart F, Suttle CA, Krisch HM . (2005). Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc Natl Acad Sci USA 102: 12471–12476.
Fuhrman JA, Caron DA . (2016). Heterotrophic Planktonic Microbes: Virus, Bacteria, Archaea, and Protozoa. In: Manual of Environmental Microbiology, Yates M, Nakatsu C, Miller R, Pillai S (eds), ASM Press: Washington, DC, USA, pp 4.2.2 –1.4.2.2 –34..
Fuhrman JA, Cram JA, Needham DM . (2015). Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol 13: 133–146.
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S . (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109.
García-Martínez J, Rodríguez-Valera F . (2000). Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of Group I. Mol Ecol 9: 935–948.
Goldsmith DB, Brum JR, Hopkins M, Carlson CA, Breitbart M . (2015). Water column stratification structures viral community composition in the Sargasso Sea. Aquat Microb Ecol 76: 85–94.
Guindon S, Gascuel O . (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704.
Holmfeldt K, Howard-Varona C, Solonenko N, Sullivan MB . (2014). Contrasting genomic patterns and infection strategies of two co-existing bacteroidetes podovirus genera. Environ Microbiol 16: 2501–2513.
Howard-Varona C, Roux S, Dore H, Solonenko NE, Holmfeldt K, Markillie LM et al. (2016). Regulation of infection efficiency in a globally abundant marine bacteriodetes virus. ISME J 11: 284–295.
Ignacio-Espinoza JC, Sullivan MB . (2012). Phylogenomics of T4 cyanophages: lateral gene transfer in the 'core' and origins of host genes. Environ Microbiol 14: 2113–2126.
Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A et al. (2014). Single-cell genomics reveals hundreds of coexisting subpopulations in wild prochlorococcus. Science 344: 416–420.
Katoh K, Misawa K, Kuma K, Miyata T . (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066.
Labonté JM, Swan BK, Poulos B, Luo H, Koren S, Hallam SJ et al. (2015). Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J 9: 2386–2399.
Lu H-P, Yeh Y-C, Sastri AR, Shiah F-K, Gong G-C, Hsieh C . (2016). Evaluating community–environment relationships along fine to broad taxonomic resolutions reveals evolutionary forces underlying community assembly. ISME J 10: 2867–2878.
Luo H, Swan BK, Stepanauskas R, Hughes AL, Moran MA . (2014). Comparing effective population sizes of dominant marine alphaproteobacteria lineages. Environ Microbiol Rep 6: 167–172.
Marston MF, Amrich CG . (2009). Recombination and microdiversity in coastal marine cyanophages. Environ Microbiol 11: 2893–2903.
Marston MF, Pierciey FJ, Shepard A, Gearin G, Qi J, Yandava C et al. (2012). Rapid diversification of coevolving marine synechococcus and a virus. Proc Natl Acad Sci USA 109: 4544–4549.
Morris RM, Rappé MS, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ . (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810.
Needham DM, Chow C-ET, Cram JA, Sachdeva R, Parada AE, Fuhrman JA . (2013). Short-term observations of marine bacterial and viral communities: patterns, connections and resilience. ISME J 7: 1274–1285.
Needham DM, Fuhrman JA . (2016). Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol 1: 16005.
Newton RJ, Mclellan SL, Dila DK, Vineis JH, Morrison HG, Eren AM et al. (2015). Sewage reflects the microbiomes of human populations. MBio 6: 1–9.
Nolan JM, Petrov V, Bertrand C, Krisch HM, Karam JD . (2006). Genetic diversity among five T4-like bacteriophages. Virol J 3: 30.
Pagarete A, Chow C-ET, Johannessen T, Fuhrman JA, Thingstad TF, Sandaa RA . (2013). Strong seasonality and interannual recurrence in marine myovirus communities. Appl Environ Microbiol 79: 6253–6259.
Parada AE, Needham DM, Fuhrman JA . (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time-series and global field samples. Environ Microbiol 18: 1403–1414.
Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD . (2010). Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J 7: 292.
Polz MF, Hunt DE, Preheim SP, Weinreich DM . (2006). Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos Trans R Soc B 361: 2009–2021.
Preheim SP, Perrott AR, Martin-Platero AM, Gupta A, Alm EJ . (2013). Distribution-based clustering: using ecology to refine the operational taxonomic unit. Appl Environ Microbiol 79: 6593–6603.
R Core Team. (2015). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Reveillaud J, Maignien L, Murat Eren A, Huber JA, Apprill A, Sogin ML et al. (2014). Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J 8: 1198–1209.
Rice P, Longden I, Bleasby A . (2000). EMBOSS: the European Molecular Biology Open Software Suite (2000). Trends Genet 16: 276–277.
Riemann L, Grossart HP . (2008). Elevated lytic phage production as a consequence of particle colonization by a marine flavobacterium (Cellulophaga sp.). Microb Ecol 56: 505–512.
Rocap G, Distel DL, Waterbury JB, Chisholm SW . (2002). Resolution of prochlorococcus and synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol 68: 1180–1191.
Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA et al. (2003). Genome divergence in two prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047.
Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M et al. (2010). Viral and microbial community dynamics in four aquatic environments. ISME J 4: 739–751.
Roger F, Godhe A, Gamfeldt L . (2012). Genetic diversity and ecosystem functioning in the face of multiple stressors. PLoS One 7: e45007.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson SJ, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504.
Sjöqvist CO, Kremp A . (2016). Genetic diversity affects ecological performance and stress response of marine diatom populations. ISME J 10: 2755–2766.
Storey JD, Tibshirani R . (2003). Statistical significance for genome wide studies. Proc Natl Acad Sci USA 100: 9440–9445.
Sullivan MB, Waterbury JB, Chisholm SW . (2003). Cyanophages infecting the oceanic cyanobacterium prochlorococcus. Nature 424: 1047–1052.
Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611.
Teeling H, Fuchs BM, Bennke CM, Krüger K, Chafee M, Kappelmann L et al. (2016). Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. Elife 5: 1–31.
Tesson SVM, Montresor M, Procaccini G, Kooistra WHCF . (2014). Temporal changes in population structure of a marine planktonic diatom. PLoS One 9: 1–23.
Thingstad TF, Våge S, Storesund JE, Sandaa R-A, Giske J . (2014). A theoretical analysis of how strain-specific viruses can control microbial species diversity. Proc Natl Acad Sci USA 111: 7813–7818.
Thompson AW, Foster RA, Krupke A, Carter BJ, Musat N, Vaulot D et al. (2012). Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337: 1546–1550.
Tikhonov M, Leach RW, Wingreen NS . (2015). Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J 9: 68–80.
Tully BJ, Nelson WC, Heidelberg JF . (2011). Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine. Environ Microbiol 14: 254–267.
Turlapati SA, Minocha R, Long S, Ramsdell J, Minocha SC . (2015). Oligotyping reveals stronger relationship of organic soil bacterial community structure with N-amendments and soil chemistry in comparison to that of mineral soil at Harvard Forest, MA, USA. Front Microbiol 6: 1–16.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Wang D, Zhang Y, Zhang Z, Zhu J, Yu J . (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8: 77–80.
Xia LC, Ai D, Cram JA, Fuhrman JA, Sun F . (2013). Efficient statistical significance approximation for local similarity analysis of high-throughput time series data. Bioinformatics 29: 230–237.
Xia LC, Steele JA, Cram JA, Cardon ZG, Simmons SL, Vallino JJ et al. (2011). Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst Biol 5 (Suppl 2): S15.
Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K et al. (2007). The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol 5: e16.
Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC et al. (2013). Abundant SAR11 viruses in the ocean. Nature 494: 357–360.
Acknowledgements
We thank the USC Wrigley Institute of Environmental Science, especially Roberta Marinelli, Sean Conner and Captain Gordon Boivin, and the crew of the Miss Christie for sampling opportunities and laboratory space. We thank Mark V Brown for his providing of SAR11 database used for classification of SAR11 ITS sequences. We thank Jennifer Chang, Cheryl Chow, Alle Lie, Sean McCallister and Elizabeth Teel for sampling assistance. We thank Dave Caron, Frank Corsetti, John Heidelberg, Eric Webb, Nathan Ahlgren, Lyria Berdjeb, Jacob Cram, Laura Gómez Consarnau, Erin Fichot, Alma Parada, Ella Sieradzki, Yi-Chun Yeh and Yuanqi Wang for insightful discussion and feedback on the manuscript. This work was supported by NSF Grants 1031743 and 1136818, grant GBMF3779 from the Gordon and Betty Moore Foundation Marine Microbiology Initiative and a National Science Foundation Graduate Student Research Fellowship to DMN.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Needham, D., Sachdeva, R. & Fuhrman, J. Ecological dynamics and co-occurrence among marine phytoplankton, bacteria and myoviruses shows microdiversity matters. ISME J 11, 1614–1629 (2017). https://doi.org/10.1038/ismej.2017.29
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.29
This article is cited by
-
Disentangling microbial networks across pelagic zones in the tropical and subtropical global ocean
Nature Communications (2024)
-
Seasonal changes in biodiversity of native and non-native amphipod taxa under diverse environmental contexts
Marine Biology (2024)
-
Assembly processes and functional diversity of marine protists and their rare biosphere
Environmental Microbiome (2023)
-
Disentangling temporal associations in marine microbial networks
Microbiome (2023)
-
Environment drives the co-occurrence of bacteria and microeukaryotes in a typical subtropical bay
Journal of Oceanology and Limnology (2023)