Abstract
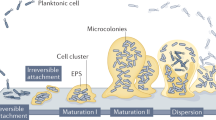
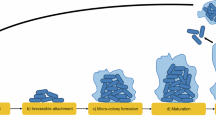
Bacteria can grow in a free-swimming state, as planktonic cells, or in surface-attached communities, termed biofilms. The planktonic and biofilm growth modes differ dramatically with respect to spatial constraints, nutrient access, population density and cell–cell interactions. Fitness trade-offs underlie how successfully bacteria compete in each of these environments. Accordingly, some bacteria have evolved to be specialists in biofilm formation, while others specialize in planktonic growth. There are species, however, that possess flexible strategies: they can transition between the molecular programs required for biofilm formation and for planktonic growth. Such flexible strategies often sacrifice competitive ability against specialists in a given habitat. There is little exploration of the ecological conditions favoring the evolution of the flexible biofilm production strategy for bacteria in competition with specialist biofilm producers or specialist non-producers. Here, we study the human pathogen Vibrio cholerae, a flexible biofilm-former, as well as constitutive biofilm-producing and non-producing mutants. We assess the fitness of these strains under biofilm conditions, planktonic conditions and conditions that demand the ability to transition between the two growth modes. We show that, relative to the specialists, the wild type is superior at dispersal from biofilms to the planktonic phase; however, this capability comes at the expense of reduced competitive fitness against constitutive biofilm producers on surfaces. Wild-type V. cholerae can outcompete the constitutive biofilm producers and non-producers if habitat turnover is sufficiently frequent. Thus, selection for phenotypic flexibility in biofilm production depends on the frequency of environmental fluctuations encountered by bacteria.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ali JG, Agrawal AA . (2012). Specialist versus generalist insect herbivores and plant defense. Trends Plant Sci 17: 293–302.
Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ et al. (2004). Global gene expression in Staphylococcus aureus biofilms. J Bacteriol 186: 4665–4684.
Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH . (2007). Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J Bacteriol 189: 388–402.
Beyhan S, Yildiz FH . (2007). Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol Microbiol 63: 995–1007.
Bilecen K, Yildiz FH . (2009). Identification of a calcium-controlled negative regulatory system affecting Vibrio cholerae biofilm formation. Environ Microbiol 11: 2015–2029.
Chowdhury G, Bhadra RK, Bag S, Pazhani GP, Das B, Basu P et al. (2016). Rugose atypical Vibrio cholerae O1 El Tor responsible for 2009 cholera outbreak in India. J Med Microbiol 65: 1130–1136.
Conner JG, Teschler JK, Jones CJ, Yildiz FH . (2016). Staying alive: Vibrio cholerae’s cycle of environmental survival, transmission, and dissemination. Microbiol Spectr 4: doi:10.1128/microbiolspec.VMBF-0015-2015.
Devictor V, Julliard R, Jiguet F . (2008). Distribution of specialist and generalist species along spatial gradients of habitat disturbance and fragmentation. Oikos 117: 507–514.
Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL . (2014). Solutions to the public goods dilemma in bacterial biofilms. Curr Biol 24: 50–55.
Drescher K, Dunkel J, Nadell CD, van Teeffelen S, Grnja I, Wingreen NS et al. (2016). Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc Natl Acad Sci USA 113: E2066–E2072.
Egas M, Dieckmann U, Sabelis MW . (2004). Evolution restricts the coexistence of specialists and generalists: the role of trade-off structure. Am Nat 163: 518–531.
Flemming H-C, Wingender J . (2010). The biofilm matrix. Nat Rev Microbiol 8: 623–633.
Fong JCN, Yildiz FH . (2007). The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol 189: 2319–2330.
Fong JCN, Syed KA, Klose KE, Yildiz FH . (2010). Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiol 156: 2757–2769.
Hall-Stoodley L, Costerton JW, Stoodley P . (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108.
Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI . (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436: 1171–1175.
Irie Y, Roberts A, Kragh KN, Gordon VD, Hutchison J, Allen RJ et al (2016). The Pseudomonas aeruginosa PSL polysaccharide is a social but non-cheatable trait in biofilms. bioRxiv: doi: https://doi.org/10.1101/049783.
Kassen R . (2002). The experimental evolution of specialists, generalists, and the maintenance of diversity. J Evol Biol 15: 173–190.
Kim W, Racimo F, Schluter J, Levy SB, Foster KR . (2014). Importance of positioning for microbial evolution. Proc Natl Acad Sci USA 111: E1639–E1647.
Kragh KN, Hutchison JB, Melaugh G, Rodesney C, Roberts AEL, Irie Y et al. (2016). Role of multicellular aggregates in biofilm formation. mBio 7: e00237.
Lowe WH, McPeek MA . (2014). Is dispersal neutral? Trends Ecol Evol 29: 444–450.
Madsen JS, Lin Y-C, Squyres GR, Price-Whelan A, de Santiago Torio A, Song A et al. (2015). Facultative control of matrix production optimizes competitive fitness in Pseudomonas aeruginosa PA14 biofilm models. Appl Environ Microbiol 81: 8414–8426.
Martínez-García E, Nikel PI, Chavarría M, Lorenzo V . (2014). The metabolic cost of flagellar motion in Pseudomonas putida KT2440. Environ Microbiol 16: 291–303.
McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S . (2012). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Micro 10: 39–50.
Meibom KL, Li XBB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK . (2004). The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci USA 101: 2524–2529.
Mikkelsen H, Duck Z, Lilley KS, Welch M . (2007). Interrelationships between colonies, biofilms, and planktonic cells of Pseudomonas aeruginosa. J Bacteriol 189: 2411–2416.
Millet YA, Alvarez D, Ringgaard S, von Andrian UH, Davis BM, Waldor MK . (2014). Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog 10: e1004405.
Müller J, Miller MC, Nielsen AT, Schoolnik GK, Spormann AM . (2007). vpsA-and luxO-independent biofilms of Vibrio cholerae. FEMS Microbiol Lett 275: 199–206.
Nadell CD, Xavier JB, Levin SA, Foster KR . (2008). The evolution of quorum sensing in bacterial biofilms. PLoS Biol 6: e14.
Nadell CD, Xavier JB, Foster KR . (2009). The sociobiology of biofilms. FEMS Microbiol Rev 33: 206–224.
Nadell CD, Bassler BL . (2011). A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci USA 108: 14181–14185.
Nadell CD, Drescher K, Wingreen NS, Bassler BL . (2015). Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J 9: 1700–1709.
Nadell CD, Drescher K, Foster KR . (2016). Spatial structure, cooperation, and competition in bacterial biofilms. Nat Rev Microbiol 14: 589–600.
Nelson EJ, Harris JB, Glenn Morris J, Calderwood SB, Camilli A . (2009). Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat Rev Microbiol 7: 693–702.
Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK . (2006). RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog 2: 933–948.
Oliveira NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W et al. (2015). Biofilm formation as a response to ecological competition. PLoS Biol 13: e1002191.
Papenfort K, Förstner KU, Cong J-P, Sharma CM, Bassler BL . (2015). Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci USA 112: E766–E775.
Schluter J, Nadell CD, Bassler BL, Foster KR . (2015). Adhesion as a weapon in microbial competition. ISME J 9: 139–149.
Sengupta C, Mukherjee O, Chowdhury R . (2016). Adherence to intestinal cells promotes biofilm formation in Vibrio cholerae. J Infect Dis 214: 1571–1578.
Skorupski K, Taylor RK . (1996). Positive selection vectors for allelic exchange. Gene 169: 47–52.
Smith DR, Maestre-Reyna M, Lee G, Gerard H, Wang AH-J, Watnick PI . (2015). In situ proteolysis of the Vibrio cholerae matrix protein RbmA promotes biofilm recruitment. Proc Natl Acad Sci USA 112: 10491–10496.
Snell-Rood EC . (2013). An overview of the evolutionary causes and consequences of behavioural plasticity. Anim Behav 85: 1004–1011.
Stewart PS, Franklin MJ . (2008). Physiological heterogeneity in biofilms. Nat Rev Microbiol 6: 199–210.
Stewart PS . (2012). Mini-review: convection around biofilms. Biofouling 28: 187–198.
Stocker R, Seymour JR, Samandani A, Hunt DE, Polz MF . (2008). Rapic chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA 105: 4209–4214.
Stocker R, Seymour JR . (2012). Ecology and physics of bacterial chemotaxis in the ocean. Microbiol Mol Biol Rev 76: 792–812.
Teschler JK, Zamorano-Sanchez D, Utada AS, Warner CJA, Wong GCL, Linington RG et al. (2015). Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol 13: 255–268.
Utada AS, Bennett RR, Fong JCN, Gibiansky ML, Yildiz FH, Golestanian R et al. (2014). Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat Commun 5: 4913.
Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S et al. (2001). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413: 860–864.
Wilson DS, Yoshimura J . (1994). On the coexistence of specialists and generalists. Am Nat 144: 692–707.
Xavier JB, Foster KR . (2007). Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA 104: 876–881.
Yan J, Sharo AG, Stone HA, Wingreen NS, Bassler BL . (2016). Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc Natl Acad Sci USA 113: E5337–E5343.
Yawata Y, Cordero OX, Menolascina F, Hehemann J-H, Polz MF, Stocker R . (2014). Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci USA 111: 5622–5627.
Acknowledgements
This work was supported by the Alexander von Humboldt Foundation (CDN), Howard Hughes Medical Institute (BLB), NIH Grant 2R37GM065859 (BLB) and National Science Foundation Grant MCB-0948112 (BLB). JY holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Author contributions
CDN and JY initiated this project. JY and CDN constructed the strains. JY performed the experiments. JY, CDN and BLB analyzed the data. CDN, JY and BLB wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Yan, J., Nadell, C. & Bassler, B. Environmental fluctuation governs selection for plasticity in biofilm production. ISME J 11, 1569–1577 (2017). https://doi.org/10.1038/ismej.2017.33
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.33
This article is cited by
-
Surface remodeling and inversion of cell-matrix interactions underlie community recognition and dispersal in Vibrio cholerae biofilms
Nature Communications (2025)
-
Vibrio cholerae biofilms use modular adhesins with glycan-targeting and nonspecific surface binding domains for colonization
Nature Communications (2023)
-
An RNA sponge controls quorum sensing dynamics and biofilm formation in Vibrio cholerae
Nature Communications (2022)
-
Bivariate genome-wide association study of the growth plasticity of Staphylococcus aureus in coculture with Escherichia coli
Applied Microbiology and Biotechnology (2020)
-
Microbial life cycles link global modularity in regulation to mosaic evolution
Nature Ecology & Evolution (2019)