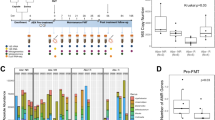
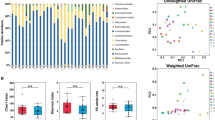
Abstract
Faecal microbiota transplantation (FMT) may contribute towards disease remission in ulcerative colitis (UC), but it is unknown which factors determine long-term effect of treatment. Here, we aimed to identify bacterial signatures associated with sustained remission. To this end, samples from healthy donors and UC patients—grouped into responders and non-responders at a primary end point (week 12) and further stratified by sustained clinical remission and relapse assessed at ⩾1-year follow-up were analysed, comparing the efficacy of FMT from either a healthy donor or autologous faeces. Microbiota composition was determined with a 16S rRNA gene-based phylogenetic microarray on faecal and mucosal samples, and functional profiles were predicted using PICRUSt with quantitative PCR verification of the butyrate production capacity; short-chain fatty acids were measured in faecal samples. At baseline, UC patients showed reduced amounts of bacterial groups from the Clostridium cluster XIVa, and significantly higher levels of Bacteroidetes as compared with donors. These differences were reduced after FMT mostly in responders. Sustained remission was associated with known butyrate producers and overall increased butyrate production capacity, while relapse was associated with Proteobacteria and Bacteroidetes. Ruminococcus gnavus was found at high levels in donors of failed FMT. A microbial ecosystem rich in Bacteroidetes and Proteobacteria and low in Clostridium clusters IV and XIVa observed in UC patients after FMT was predictive of poor sustained response, unless modified with a donor microbiota rich in specific members from the Clostridium clusters IV and XIVa. Additionally, sustained response was associated with restoration of the butyrate production capacity.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Angelberger S, Lichtenberger C, Gratzer C, Papay P, Primas C, Eser A et al. (2012). P374 Fecal transplantation in patients with moderately to severely chronic active ulcerative colitis (UC). J Crohn's Colitis 6: S159–S159.
Aroniadis OC, Brandt LJ, Greenberg A, Borody T, Kelly CR, Mellow M et al. (2016). Long-term follow-up study of fecal microbiota transplantation for severe and/or complicated Clostridium difficile infection: a multicenter experience. J Clin Gastroenterol 50: 398–402.
Basset C, Holton J, Bazeos A, Vaira D, Bloom S . (2004). Are Helicobacter species and enterotoxigenic Bacteroides fragilis involved in inflammatory bowel disease? Dig Dis Sci 49: 1425–1432.
Benjamini Y, Hochberg Y . (1995). Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300.
Berry D, Reinisch W . (2013). Intestinal microbiota: a source of novel biomarkers in inflammatory bowel diseases? Best Pract Res Clin Gastroenterol 27: 47–58.
Boesmans L, Ramakers M, Arijs I, Windey K, Vanhove W, Schuit F et al. (2015). Inflammation-induced downregulation of butyrate uptake and oxidation is not caused by a reduced gene expression. J Cell Physiol 230: 418–426.
Borody T, Wettstein A, Campbell J, Leis S, Torres M, Finlayson S et al. (2012). Fecal microbiota transplantation in ulcerative colitis: review of 24 years experience. Am J Gastroenterol 107: S665–S665.
Claesson MJ, O'Sullivan O, Wang Q, Nikkila J, Marchesi JR, Smidt H et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4: e6669.
De Preter V, Machiels K, Joossens M, Arijs I, Matthys C, Vermeire S et al. (2015). Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut 64: 447–458.
Dicksved J, Halfvarson J, Rosenquist M, Jarnerot G, Tysk C, Apajalahti J et al. (2008). Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J 2: 716–727.
Eeckhaut V, Machiels K, Perrier C, Romero C, Maes S, Flahou B et al. (2013). Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 62: 1745–1752.
Frank DN St, Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR . (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785.
Huda-Faujan N, Abdulamir AS, Fatimah AB, Anas OM, Shuhaimi M, Yazid AM et al. (2010). The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem J 4: 53–58.
Iraporda C, Errea A, Romanin DE, Cayet D, Pereyra E, Pignataro O et al. (2015). Lactate and short chain fatty acids produced by microbial fermentation downregulate proinflammatory responses in intestinal epithelial cells and myeloid cells. Immunobiology 220: 1161–1169.
Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P et al. (2011). Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 60: 631–637.
Jowett SL, Seal CJ, Phillips E, Gregory W, Barton JR, Welfare MR . (2003). Defining relapse of ulcerative colitis using a symptom-based activity index. Scand J Gastroenterol 38: 164–171.
Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M et al. (2010). Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis 16: 2034–2042.
Kaser A, Zeissig S, Blumberg RS . (2010). Inflammatory bowel disease. Annu Rev Immunol 28: 573–621.
Kostic AD, Xavier RJ, Gevers D . (2014). The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146: 1489–1499.
Kump PK, Grochenig HP, Lackner S, Trajanoski S, Reicht G, Hoffmann KM et al. (2013). Alteration of intestinal dysbiosis by fecal microbiota transplantation does not induce remission in patients with chronic active ulcerative colitis. Inflamm Bowel Dis 19: 2155–2165.
Kunde S, Pham A, Bonczyk S, Crumb T, Duba M, Conrad H Jr et al. (2013). Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J Pediatr Gastroenterol Nutr 56: 597–601.
Lahti L, Salojarvi J, Salonen A, Scheffer M, de Vos WM . (2014). Tipping elements in the human intestinal ecosystem. Nat Commun 5: 4344.
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821.
Louis P, Flint HJ . (2007). Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol 73: 2009–2012.
Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V et al. (2014). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63: 1275–1283.
Maharshak N, Huh EY, Paiboonrungruang C, Shanahan M, Thurlow L, Herzog J et al. (2015). Enterococcus faecalis gelatinase mediates intestinal permeability via protease-activated receptor 2. Infect Immun 83: 2762–2770.
Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F et al. (2007). Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res 6: 546–551.
McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D et al. (2012). The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience 1: 7.
Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C et al. (2015). Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149: 102–109 e106.
Nadkarni MA, Martin FE, Jacques NA, Hunter N . (2002). Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148: 257–266.
Nagalingam NA, Lynch SV . (2012). Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 18: 968–984.
Ohkusa T, Yoshida T, Sato N, Watanabe S, Tajiri H, Okayasu I . (2009). Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitis. J Med Microbiol 58: 535–545.
Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR et al. (2004). Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53: 685–693.
Paramsothy S, Kamm M, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D et al. (2017). Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389: 1218–1228.
Petersen AM, Halkjaer SI, Gluud LL . (2015). Intestinal colonization with phylogenetic group B2 Escherichia coli related to inflammatory bowel disease: a systematic review and meta-analysis. Scand J Gastroenterol 50: 1–9.
Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI et al. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105: 2420–2428.
Rajilic-Stojanovic M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H et al. (2009). Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol 11: 1736–1751.
Rajilic-Stojanovic M, Shanahan F, Guarner F, de Vos WM . (2013). Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis 19: 481–488.
Rajilić-Stojanović M, Heilig HGHJ, Tims S, Zoetendal EG, de Vos WM . (2013). Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol 15: 1146–1159.
Rangel I, Sundin J, Fuentes S, Repsilber D, de Vos WM, Brummer RJ . (2015). The relationship between faecal-associated and mucosal-associated microbiota in irritable bowel syndrome patients and healthy subjects. Aliment Pharmacol Ther 42: 1211–1221.
Rossen NG, Fuentes S, van der Spek MJ, Tijssen JG, Hartman JH, Duflou A et al. (2015). Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 149: 110–118 e114.
Salonen A, Nikkila J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, Kekkonen RA et al. (2010). Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods 81: 127–134.
Sartor RB . (2006). Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol 3: 390–407.
Stams AJ, Van Dijk JB, Dijkema C, Plugge CM . (1993). Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl Environ Microbiol 59: 1114–1119.
Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L . (2014). The role of short-chain fatty acids in health and disease. Adv Immunol 121: 91–119.
ter Braak CJF, Šmilauer P . (2012) Canoco Reference Manual and User's Guide: Software For Ordination, Version 5.0. Microcomputer Power: Ithaca, NY, USA.
van den Bogert B, de Vos WM, Zoetendal EG, Kleerebezem M . (2011). Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol 77: 2071–2080.
Vermeiren J, Van den Abbeele P, Laukens D, Vigsnaes LK, De Vos M, Boon N et al. (2012). Decreased colonization of fecal Clostridium coccoides/Eubacterium rectale species from ulcerative colitis patients in an in vitro dynamic gut model with mucin environment. FEMS Microbiol Ecol 79: 685–696.
Vernia P, Annese V, Bresci G, d'Albasio G, D'Inca R, Giaccari S et al. (2003). Topical butyrate improves efficacy of 5-ASA in refractory distal ulcerative colitis: results of a multicentre trial. Eur J Clin Invest 33: 244–248.
Wexler HM . (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20: 593–621.
Winter SE, Lopez CA, Baumler AJ . (2013). The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 14: 319–327.
Zoetendal EG, Heilig HG, Klaassens ES, Booijink CC, Kleerebezem M, Smidt H et al. (2006). Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protocols 1: 870–873.
Acknowledgements
We thank all laboratory technicians who performed HITChip, the trial nurses involved in the TURN trial and all participants who either received or donated faeces for FMT treatments. This work was supported by MLDS Grant 2011 (WO 11-17) to NGR and NWO-Spinoza Grant 2008 to WMdV. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author contributions
CP was responsible for the study concept and design. The original study protocol was written by NR and CP. NR and CP predefined end points of the trial. CP and WdV obtained funding. NR was the executive investigator of the trial and MvdS was involved in the clinical work as a trial nurse. WdV, EZ, JH and SF were responsible for all microbiological experiments. LH and KK performed qPCR experiments on butyrate production capacity. PICRUSt experiments on functional capacity of the microbiota were carried out by JS. NR and SF performed statistical analyses. SF, NR, EZ, CP and GD drafted the manuscript. Critical revision of the manuscript was done by all coauthors. All authors had access to the study data and have reviewed and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Fuentes, S., Rossen, N., van der Spek, M. et al. Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. ISME J 11, 1877–1889 (2017). https://doi.org/10.1038/ismej.2017.44
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.44
This article is cited by
-
Multi-aspect therapeutic effects of a polyphenolic herbal formulation Cirsium japonicum, Scutellaria baicalensis, Paeonia japonica, and Glycyrrhiza uralensis on ulcerative colitis: inflammation modulation, gut microbiota remodeling, and metabolite profiling
Gut Pathogens (2026)
-
Gentiopicroside improves DSS-induced ulcerative colitis and secondary liver injury in mice by enhancing the intestinal barrier and regulating the gut microbiome
Scientific Reports (2025)
-
Reduced gut microbiota diversity in ulcerative colitis patients with latent tuberculosis infection during vedolizumab therapy: insights on prophylactic anti-tuberculosis effects
BMC Microbiology (2024)
-
Human-derived bacterial strains mitigate colitis via modulating gut microbiota and repairing intestinal barrier function in mice
BMC Microbiology (2024)
-
The development of probiotics and prebiotics therapy to ulcerative colitis: a therapy that has gained considerable momentum
Cell Communication and Signaling (2024)