Abstract
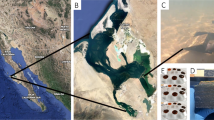
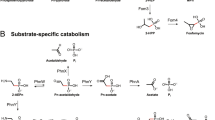
Metatranscriptomics and metagenomics data sets benchmarked with internal standards were used to characterize the expression patterns for biogeochemically relevant bacterial and archaeal genes mediating carbon, nitrogen, phosphorus and sulfur uptake and metabolism through the salinity gradient of the Amazon River Plume. The genes were identified in 48 metatranscriptomic and metagenomic data sets summing to >500 million quality-controlled reads from six locations in the plume ecosystem. The ratio of transcripts per gene copy (a direct measure of expression made possible by internal standard additions) showed that the free-living bacteria and archaea exhibited only small changes in the expression levels of biogeochemically relevant genes through the salinity and nutrient zones of the plume. In contrast, the expression levels of genes in particle-associated cells varied over orders of magnitude among the stations, with the largest differences measured for genes mediating aspects of nitrogen cycling (nifH, amtB and amoA) and phosphorus acquisition (pstC, phoX and phoU). Taxa varied in their baseline gene expression levels and extent of regulation, and most of the spatial variation in the expression level could be attributed to changes in gene regulation after removing the effect of shifting taxonomic composition. We hypothesize that changes in microbial element cycling along the Amazon River Plume are largely driven by shifting activities of particle-associated cells, with most activities peaking in the mesohaline regions where N2 fixation rates are elevated.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Amon R, Benner R . (1998). Seasonal patterns of bacterial abundance and production in the Mississippi River plume and their importance for the fate of enhanced primary production. Microb Ecol 35: 289–300.
Beier S, Rivers AR, Moran MA, Obernosterer I . (2015). The transcriptional response of prokaryotes to phytoplankton‐derived dissolved organic matter in seawater. Environ Microbiol 17: 3466–3480.
Church MJ, Wai B, Karl DM, DeLong EF . (2010). Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol 12: 679–688.
Coles VJ, Brooks MT, Hopkins J, Stukel MR, Yager PL, Hood RR . (2013). The pathways and properties of the Amazon River Plume in the tropical North Atlantic Ocean. J Geophys Res-Oceans 118: 6894–6913.
Cooley SR, Coles VJ, Subramaniam A, Yager PL . (2007). Seasonal variations in the Amazon Plume-related atmospheric carbon sink. Glob Biogeochem Cycles 21: GB3014.
Cottrell MT, Kirchman DL . (2016). Transcriptional control in marine copiotrophic and oligotrophic bacteria with streamlined genomes. Appl Environ Microb 82: 6010–6018.
Crump BC, Baross JA . (2000). Characterization of the bacterially-active particle fraction in the Columbia River estuary. Mar Ecol Prog Ser 206: 13–22.
Dagg M, Benner R, Lohrenz S, Lawrence D . (2004). Transformation of dissolved and particulate materials on continental shelves influenced by large rivers: plume processes. Continental Shelf Res 24: 833–858.
Dagg MJ, Breed GA . (2003). Biological effects of Mississippi River nitrogen on the northern Gulf of Mexico—a review and synthesis. J Mar Syst 43: 133–152.
Foster RA, Kuypers MM, Vagner T, Paerl RW, Musat N, Zehr JP . (2011). Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J 5: 1484–1493.
Gardner WS, Cotner JB, Eadie BJ, Cavaletto JF, Benner R, Chin-Leo G . (1994). Mineralization of organic material and bacterial dynamics in Mississippi River plume water. Estuaries 17: 816–828.
Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA . (2011). Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J 5: 461–472.
Gifford SM, Becker JW, Sosa OA, Repeta DJ, DeLong EF . (2016). Quantitative transcriptomics reveals the growth- and nutrient-dependent response of a streamlined marine methylotroph to methanol and naturally occurring dissolved organic matter. mBio 7: e01279–e01216.
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Goebel NL, Turk KA, Achilles KM, Paerl R, Hewson I, Morrison AE et al. (2010). Abundance and distribution of major groups of diazotrophic cyanobacteria and their potential contribution to N2 fixation in the tropical Atlantic Ocean. Environ Microbiol 12: 3272–3289.
Goes JI, Gomes HdR, Chekalyuk AM, Carpenter EJ, Montoya JP, Coles VJ et al. (2014). Influence of the Amazon River discharge on the biogeography of phytoplankton communities in the western tropical north Atlantic. Prog Oceanogr 120: 29–40.
Hilton JA, Satinsky BM, Doherty M, Zielinski BL, Zehr JP . (2014). Metatranscriptomics of N2-fixing cyanobacteria in the Amazon River plume. ISME J 9: 1557–1569.
Howard EC, Henriksen JR, Buchan A, Reisch CR, Bürgmann H, Welsh R et al. (2006). Bacterial taxa that limit sulfur flux from the ocean. Science 314: 649–652.
Huang Y, Niu B, Gao Y, Fu L, Li W . (2010). CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26: 680–682.
Kellogg CT, Carpenter SD, Renfro AA, Sallon A, Michel C, Cochran JK et al. (2011). Evidence for microbial attenuation of particle flux in the Amundsen Gulf and Beaufort Sea: elevated hydrolytic enzyme activity on sinking aggregates. Polar Biol 34: 2007–2023.
Kramer JG, Singleton FL . (1992). Variations in rRNA content of marine Vibrio spp during starvation-survival and recovery. Appl Environ Microb 58: 201–207.
Lankiewicz TS, Cottrell MT, Kirchman DL . (2016). Growth rates and rRNA content of four marine bacteria in pure cultures and in the Delaware estuary. ISME J 10: 823–832.
Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S et al. (2009). The genomic basis of trophic strategy in marine bacteria. P Natl Acad Sci USA 106: 15527–15533.
Loick-Wilde N, Weber SC, Conroy BJ, Capone DG, Coles VJ, Medeiros PM et al. (2015). Nitrogen sources and net growth efficiency of zooplankton in three Amazon River plume food webs. Limnol Oceanogr 61: 460–481.
Luo H, Moran MA . (2015). How do divergent ecological strategies emerge among marine bacterioplankton lineages? Trends Microbiol 23: 577–584.
Maaloe O, Ingraham G, Maaloe O, Neidhardt F . (1983) Growth of the Bacterial Cell. Sinauer Association, Inc.: Sunderland, MA, USA.
Maier T, Schmidt A, Güell M, Kühner S, Gavin AC, Aebersold R et al. (2011). Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Sys Biol 7: 511.
McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR et al. (2010). Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci 107: 16420–16427.
Medeiros PM, Seidel M, Ward ND, Carpenter EJ, Gomes HR, Niggemann J et al. (2015). Fate of the Amazon River dissolved organic matter in the tropical Atlantic Ocean. Glob Biogeochem Cycles 29: 677–690.
Moran MA, Satinsky B, Gifford SM, Luo HW, Rivers A, Chan LK et al. (2013). Sizing up metatranscriptomics. ISME J 7: 237–243.
Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwihare LI, Buchan A et al. (2016). Deciphering ocean carbon in a changing world. Proc Natl Acad Sci USA 113: 3143–3151.
Neidhardt F, Umbarger H . (1996). Chemical composition of Escherichia coli. In: Neidhardt FC ed Escherichia coli and Salmonella: Cellular and Molecular Biology. American Society for Microbiology: Washington, DC, pp 13–16.
Pedrós-Alió C . (2006). Marine microbial diversity: can it be determined? Trends Microbiol 14: 257–263.
Polz MF, Hunt DE, Preheim SP, Weinreich DM . (2006). Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos Trans R Soc B 361: 2009–2021.
Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ et al. (2011). Detecting novel associations in large data sets. Science 334: 1518–1524.
Richey JE, Nobre C, Deser C . (1989). Amazon Rriver discharge and climate variability: 1903 to 1985. Science 246: 101–103.
Satinsky BM, Gifford SM, Crump BC, Moran MA . (2013). Use of internal standards for quantitative metatranscriptome and metagenome analysis. Methods Enzymol 531: 237–250.
Satinsky BM, Crump BC, Smith CB, Sharma S, Zielinski BL, Doherty M et al. (2014a). Microspatial gene expression patterns in the Amazon River Plume. Proc Natl Acad Sci 111: 11085–11090.
Satinsky BM, Zielinski BL, Doherty M, Smith CB, Sharma S, Paul JH et al. (2014b). The Amazon continuum dataset: quantitative metagenomic and metatranscriptomic inventories of the Amazon River plume, June 2010. Microbiome 2: 17.
Simon N, Cras AL, Foulon E, Lemée R . (2009). Diversity and evolution of marine phytoplankton. Comptes Rendus Biol 332: 159–170.
Smets W, Leff JW, Bradford MA, McCulley RL, Lebeer S, Fierer N . (2016). A method for simultaneous measurement of soil bacterial abundances and community composition via 16S rRNA gene sequencing. Soil Biol Biochem 96: 145–151.
Smith DP, Thrash JC, Nicora CD, Lipton MS, Burnum-Johnson KE, Carini P et al. (2013). Proteomic and transcriptomic analyses of Candidatus Pelagibacter ubique describe the first PII-independent response to nitrogen limitation in a free-living Alphaproteobacterium. mBio 4: e00133–e00112.
Smith WO, Demaster DJ . (1996). Phytoplankton biomass and productivity in the Amazon River plume: correlation with seasonal river discharge. Continental Shelf Res 16: 291–319.
Steglich C, Lindell D, Futschik M, Rector T, Steen R, Chisholm SW . (2010). Short RNA half-lives in the slow-growing marine cyanobacterium Prochlorococcus. Genome Biol 11: 1.
Subramaniam A, Yager PL, Carpenter EJ, Mahaffey C, Björkman K, Cooley S et al. (2008). Amazon River enhances diazotrophy and carbon sequestration in the tropical North Atlantic Ocean. Proc Natl Acad Sci 105: 10460–10465.
Taniguchi Y, Choi PJ, Li GW, Chen HY, Babu M, Hearn J et al. (2010). Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329: 533–538.
Varaljay VA, Robidart J, Preston CM, Gifford SM, Durham BP, Burns AS et al. (2015). Single-taxon field measurements of bacterial gene regulation controlling DMSP fate. ISME J 9: 1677–1686.
Yeung LY, Berelson WM, Young ED, Prokopenko MG, Rollins N, Coles VJ et al. (2012). Impact of diatom-diazotroph associations on carbon export in the Amazon River plume. Geophys Res Lett 39: L18609.
Zhao Y, Tang H, Ye Y . (2012). RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28: 125–126.
Zielinski BL, Allen AE, Carpenter EJ, Coles VJ, Crump BC, Doherty M et al. (2016). Patterns of transcript abundance of eukaryotic biogeochemically-relevant genes in the Amazon River Plume. PLoS One 11: e0160929.
Acknowledgements
We appreciate advice and assistance from Jan Mrazek, Adrian Burd, Roger Nilsen and Karie Sines and the captain and crew of the RV Knorr KN-197-08. Resources were provided by the University of Georgia’s Georgia Advanced Computing Resource Center. This work was funded by the Gordon and Betty Moore Foundation through grants GBMF2293, GBMF2928 and GBMF538.01.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Satinsky, B., Smith, C., Sharma, S. et al. Expression patterns of elemental cycling genes in the Amazon River Plume. ISME J 11, 1852–1864 (2017). https://doi.org/10.1038/ismej.2017.46
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.46
This article is cited by
-
Synechococcus nitrogen gene loss in iron-limited ocean regions
ISME Communications (2023)
-
Distinct taxonomic and ecological functions of microbiome in sediments of different depth in Bohai Sea and Yellow Sea
Journal of Oceanology and Limnology (2023)
-
Uncovering the genomic potential of the Amazon River microbiome to degrade rainforest organic matter
Microbiome (2020)