Abstract
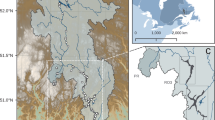
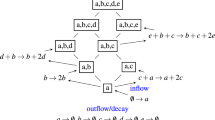
Seed banks are believed to contribute to compositional changes within and across microbial assemblages, but the application of this concept to natural communities remains challenging. Here we describe the core seed bank of a bacterial metacommunity from a boreal watershed, using the spatial distribution of bacterial operational taxonomic units (OTUs) across 223 heterogeneous terrestrial, aquatic and phyllosphere bacterial assemblages. Taxa were considered potential seeds if they transitioned from rare to abundant somewhere within the metacommunity and if they were ubiquitous and able to persist under unfavorable conditions, the latter assessed by checking their presence in three deeply sequenced samples (one soil, one river and one lake, 2.2–3 million reads per sample). We show that only a small fraction (13%) of all detected OTUs constitute a metacommunity seed bank that is shared between all terrestrial and aquatic communities, but not by phyllosphere assemblages, which seem to recruit from a different taxa pool. Our results suggest directional recruitment driven by the flow of water in the landscape, since most aquatic sequences were associated to OTUs found in a single deeply-sequenced soil sample, but only 45% of terrestrial sequences belonged to OTUs found in the two deeply-sequenced aquatic communities. Finally, we hypothesize that extreme rarity, and its interplay with water residence time and growth rates, may further constrain the size of the potential seed bank.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Aanderud ZT, Jones SE, Fierer N, Lennon JF . (2015). Resuscitation of the rare biosphere contributes to pulses of ecosystem activity. Front Microbiol 6: 24.
Aanderud ZT, Vert JC, Lennon JT, Magnusson TW, Breakwell DP, Harker AR . (2016). Bacterial dormancy is more prevalent in freshwater than in hypersaline lakes. Front Microbiol 7: 853.
Baselga A, Orme CDL . (2012). Betapart: an R package for the study of beta diversity. Methods Ecol Evol 3: 808–812.
Campbell BJ, Yua L, Heidelberg JF, Kirchman DL . (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N et al. (2012a). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6: 1621–1624.
Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA . (2012b). The Western English Channel contains a persistent microbial seed bank. ISME J 6: 1089–1093.
Clarke KR . (1993). Non-parametric multivariate analysis of changes in community structure. Aust J Ecol 18: 117–143.
Comte J, Lindström ES, Eiler A, Langenheder S . (2014). Can marine bacteria be recruited from freshwater sources and the air? ISME J 8: 2423–2430.
Crump BC, Adams HE, Hobbie JE, Kling GW . (2007). Biogeography of bacterioplankton in lakes and streams of an Arctic tundra catchment. Ecology 88: 1365–1378.
Crump BC, Amaral-Zettler LA, Kling GW . (2012). Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J 6: 1629–1639.
del Giorgio PA, Gasol JM . (2008) Physiological structure and single-cell activity in marine bacterioplankton. In: Kirchman D (ed), Microbial Ecology of the Ocean. John Wiley Sons, Inc: Hoboken, NJ, USA, pp 243–298.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R . (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200.
Galand PE, Casamayor EO, Kirchman DL, Lovejoy C . (2009). Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA 106: 22427–22432.
Gibbons SM, Caporaso JG, Pirrung M, Field D, Knight R, Gilbert JA . (2013). Evidence for a persistent microbial seed bank throughout the global ocean. Proc Natl Acad Sci USA 110: 4651–4655.
Hugoni M, Taiba N, Debroas D, Domaizon I, Doufornel IJ, Bronner G et al. (2013). Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA 110: 6004–6009.
Jones SE, Lennon JT . (2010). Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA 107: 5881–5886.
Kembel SW, O'Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL . (2014). Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci USA 111: 13715–13720.
Langenheder S, Comte J, Langenheder S, Comte J, Zha Y, Samad MS et al. (2016). Remnants of marine bacterial communities can be retrieved from deep sediments in lakes of marine origin. Environ Microbiol Rep 8: 479–485.
Lee JE, Buckley HL, Etienne RS, Lear G . (2013). Both species sorting and neutral processes drive assembly of bacterial communities in aquatic microcosms. Fems Microbiol Ecol 86: 288–302.
Lennon JT, Jones SE . (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 119: 119–130.
Lindh MV, Sjöstedt J, Andersson AF, Baltar F, Hugerth LW, Lundin D et al. (2015). Disentangling seasonal bacterioplankton population dynamics by high-frequency sampling. Environ Microbiol 17: 2459–2476.
Lindström ES, Forslund M, Algesten G, Bergström A . (2006). External control of bacterial community structure in lakes. Limnol Oceanogr 51: 339–342.
Liu L, Yang J, Yu Z, Wilkinson DM . (2015). The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J 9: 2068–2077.
Logares R, Audic S, Bass D, Bittner L, Boutte C, Christen R et al. (2014). Patterns of rare and abundant marine microbial eukaryotes. Curr Biol 24: 813–821.
Logares R, Lindström ES, Langenheder S, Logue JB, Paterson H, Laybourn-Parry J et al. (2013). Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J 7: 937–948.
Lynch MDJ, Neufeld JD . (2015). Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13: 217–229.
Magoc T, Salzberg S . (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963.
Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M . (2014). Swarm: robust and fast clustering method for amplicon-based studies. Peer J 2: e593.
Martiny JBH . (2015). Dispersal and the Microbiome: Learning how fast and how far microorganisms move will help us better understand the diversity of microbial communities. Microbe 10: 191–196.
Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK et al. (2011). Global patterns in the biogeography of bacterial taxa. Environ Microbiol 13: 135–144.
Neuenschwander SM, Pernthaler J, Posch T, Salcher MM . (2015). Seasonal growth potential of rare lake water bacteria suggest their disproportional contribution to carbon fluxes. Environ Microbiol 17: 781–795.
Niño-García JP, Ruiz-González C, del Giorgio PA . (2016a). Interactions between hydrology and water chemistry shape bacterioplankton biogeography across boreal freshwater networks. ISME J 10: 1755–1766.
Niño-García JP, Ruiz-González C, del Giorgio PA . (2016b). Landscape-scale spatial abundance distributions discriminate core from random components of lake bacterioplankton. Ecol Lett 19: 1506–1515.
Niño-García JP, Ruiz-González C, del Giorgio PA . (2017). Exploring the ecological coherence between the spatial and temporal patterns of bacterioplankton in boreal Lakes. Front Microbiol 8: 636.
Oksanen JF, Blanchet G, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2015). Vegan: community ecology package. R Package Version 2: 3–0.
Pedrós-Alió C . (2012). The rare bacterial biosphere. Ann Rev Mar Sci 4: 449–466.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Ruiz-González C, Niño-García JP, del Giorgio PA . (2015). Terrestrial origin of bacterial communities in complex boreal freshwater networks. Ecol Lett 18: 1198–1206.
R Core Team. (2013) R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing Vienna, Austria, Available from http://www.R-project.org/.
Salazar G, Cornejo-Castillo FM, Benitez-Barrios V, Fraile-Nuez E, Álvarez-SalgadonXA, Duarte CM et al. (2015). Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J 10: 596–608.
Saunders AM, Albertsen M, Vollertsen J, Nielsen PH . (2016). The activated sludge ecosystem contains a core community of abundant organisms. ISME J 10: 11–20.
Shade A, Jones SE, Caporaso JG, Handelsman J, Knight R, Fierer N et al. (2014). Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 5: e01371–01314.
Sjöstedt J, Koch-Schmidt P, Pontarp M, Canbäck B, Tunlid A, Lundberg P et al. (2012). Recruitment of members from the rare biosphere of marine bacterioplankton communities after an environmental disturbance. Appl Environ Microbiol 78: 1361–1369.
Staley C, Unno T, Gould TJ, Jarvis B, Phillips JB, Cotner JB et al. (2013). Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J Appl Microbiol 115: 1147–1158.
Valter de Oliveira LF, Margis R . (2015). The source of the river as a nursery for microbial diversity. PLoS One 10: e0120608.
Vergin KL, Done B, Carlson CA, Giovannoni SJ . (2013). Spatiotemporal distributions of rare bacterioplankton populations indicate adaptive strategies in the oligotrophic ocean. ISME J 71: 1–13.
Acknowledgements
We thank Terhi Rasilo, Annick St-Pierre, Alice Parkes, Isabelle Laforest-Lapointe, Travis Dawson, and the whole CarBBAS team for their contribution to the field and laboratory components of this research. This study is part of the program of the Carbon Biogeochemistry in Boreal Aquatic Systems (CarBBAS) Industrial Research Chair, co-funded by the Natural Science and Engineering Research Council of Canada (NSERC) and Hydro-Quebec. We acknowledge funding from FRQNT, NSERC, and the Canada Research Chairs program. CRG benefited of a Beatriu de Pinós post-doctoral fellowship (2014 BP_B 00116) from the Marie-Curie COFUND programme and the Catalan Government.
Author contributions
PdG, CRG designed the sampling, JPNG, CRG collected the data, CRG, JPNG and SK analyzed the data, CRG, JPNG, SK and CRG discussed and interpreted the results and CRG, PdG and SK wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Ruiz-González, C., Niño-García, J., Kembel, S. et al. Identifying the core seed bank of a complex boreal bacterial metacommunity. ISME J 11, 2012–2021 (2017). https://doi.org/10.1038/ismej.2017.67
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.67
This article is cited by
-
Terrestrial landscape as the main driver of bacterioplankton communities along a hydrological network located in the Colombian Amazon
Scientific Reports (2025)
-
The seed microbiota from an application perspective: an underexplored frontier in plant–microbe interactions
Crop Health (2025)
-
Terrestrial connectivity, upstream aquatic history and seasonality shape bacterial community assembly within a large boreal aquatic network
The ISME Journal (2022)
-
Soil microbial inoculation during flood events shapes headwater stream microbial communities and diversity
Microbial Ecology (2021)