Abstract
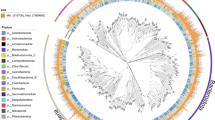
The terrestrial deep subsurface is a huge repository of microbial biomass, but in relation to its size and physical heterogeneity, few sites have been investigated in detail. Here, we applied a culture-independent metagenomic approach to characterize the microbial community composition in deep (1500 meters below surface) terrestrial fluids. Samples were collected from a former gold mine in Lead, South Dakota, USA, now Sanford Underground Research Facility (SURF). We reconstructed 74 genomes from metagenomes (MAGs), enabling the identification of common metabolic pathways. Sulfate and nitrate/nitrite reduction were the most common putative energy metabolisms. Complete pathways for autotrophic carbon fixation were found in more than half of the MAGs, with the reductive acetyl-CoA pathway by far the most common. Nearly 40% (29 of 74) of the recovered MAGs belong to bacterial phyla without any cultivated members—microbial dark matter. Three of our MAGs constitute two novel phyla previously only identified in 16 S rRNA gene surveys. The uniqueness of this data set—its physical depth in the terrestrial subsurface, the relative abundance and completeness of microbial dark matter genomes and the overall diversity of this physically deep, dark, community—make it an invaluable addition to our knowledge of deep subsurface microbial ecology.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ et al (2014). Binning metagenomic contigs by coverage and composition. Nat Methods 11: 1144–1146.
Aüllo T, Ranchou-Peyruse A, Ollivier B, Magot M (2013). Desulfotomaculum spp. and related gram-positive sulfate-reducing bacteria in deep subsurface environments. Front Microbiol 4: 362.
Baker BJ, Moser DP, MacGregor BJ, Fishbain S, Wagner M, Fry NK et al (2003). Related assemblages of sulphate‐reducing bacteria associated with ultradeep gold mines of South Africa and deep basalt aquifers of Washington State. Environ Microbiol 5: 267–277.
Baker BJ, Lazar CS, Teske AP, Dick GJ (2015). Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3: 14.
Baker BJ, Saw JH, Lind AE, Lazar CS, Hinrichs K-U, Teske AP et al (2016). Genomic inference of the metabolism of cosmopolitan subsurface Archaea, Hadesarchaea. Nat Microbiol 1: 16002.
Beal EJ, House CJ, Orphan VJ (2009). Manganese- and iron-dependent marine methane oxidation. Science 325: 184–187.
Berg IA (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77: 1925–1936.
Bolger AM, Lohse M, Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120.
Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Schulz F, Doud D et al (2017). Genome standards for single amplified genomes and genomes from metagenomes of Bacteria and Archaea. Nat Biotechnol in press.
Campbell JH, O’Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T et al (2013). UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci USA 110: 5540–5545.
Castelle CJ, Hug LA, Wrighton KC, Thomas BC, Williams KH, Wu D et al (2013). Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat Commun 4: 2120.
Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ (2015). Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol 25: 690–701.
Chivian D, Brodie EL, Alm EJ, Culley DE (2008). Environmental genomics reveals a single- species ecosystem deep within Earth. Science 322: 275–278.
Cowen JP, Giovannoni SJ, Kenig F, Johnson HP, Butterfield D, Rappé MS et al (2003). Fluids from aging ocean crust that support microbial life. Science 299: 120–123.
Creevey CJ, Doerks T, Fitzpatrick DA, Raes J, Bork P (2011). Universally distributed single-copy genes indicate a constant rate of horizontal transfer. PLoS One 6: e22099.
Darling AE, Jospin G, Lowe E, Matsen FA IV, Bik HM, Eisen JA (2014). PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 9: e243.
Derakshani M, Lukow T, Liesack W (2001). Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16 S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl Environ Microbiol 67: 623–631.
Grimm F, Franz B, Dahl C (2008)Thiosulfate and sulfur oxidation in purple sulfur bacteriaIn Microbial Sulfur Metabolism. Springer: Berlin, Heidelberg, 101–116.
Dong Y, Kumar CG, Chia N, Kim PJ, Miller PA, Price ND et al (2014). Halomonas sulfidaeris‐ dominated microbial community inhabits a 1.8 km‐deep subsurface Cambrian Sandstone reservoir. Environ Microbiol 16: 1695–1708.
Dunham JP, Friesen ML (2013). A cost-effective method for high-throughput construction of Illumina sequencing libraries. Cold Spring Harbor Protocols 2013: 820–834.
Dupont C, Rusch DB, Yooseph S, Lombardo MJ, Richter RA, Valas R et al (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6: 1186–1199.
Eddy SR (2011). Accelerated profile HMM searches. PLoS Comput Biol 10: e1002195.
Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM et al (2006). Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7: 57.
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML et al (2015). Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3: e1319.
Fredrickson JK, Onstott TC (1996). Microbes deep inside the earth. Sci Am 275: 68–73.
Ghosh W, Dam B (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol Re 33: 999–1043.
Hanson TE, Tabita FR (2001). A ribulose-1, 5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc Natl Acad Sci 8: 4397–4402.
Hugenholtz P, Pitulle C, Hershberger KL, Pace NR (1998). Novel division level bacterial diversity in a Yellowstone hot spring. Journal of Bacteriology 180: 366–376.
Hügler M, Sievert SM (2010). Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Marine Sci 3: 261–289.
Huntemann M, Ivanova NN, Mavromatis K, Tripp HJ, Paez-Espino D, Palaniappan K et al (2015). The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v. 4). Stand Genomic Sci 10: 86.
Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC (2012). Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28: 2223–2230.
Jungbluth SP, Grote J, Lin HT, Cowen JP, Rappé MS (2013). Microbial diversity within basement fluids of the sediment-buried Juan de Fuca Ridge flank. ISME J 7: 161–172.
Jungbluth SP, Glavina del Rio T, Tringe SG, Stepanauskas R, Rappé MS (2017). Genomic comparisons of a bacterial lineage that inhabits both marine and terrestrial deep subsurface systems. PeerJ 5: e3134.
Kallmeyer J, Pockalny R, Adhikari RR (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA 109: 16213–16216.
Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ et al (2013). Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio 4: e708–e713.
Kolinko S, Richter M, Glöckner FO, Brachmann A, Schüler D (2015). Single‐cell genomics of uncultivated deep‐branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ Microbiol 18: 21–37.
Langmead B, Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359.
Lau MCY, Cameron C, Magnabosco C, Brown CT, Schilkey F, Grim S et al (2014). Phylogeny and phylogeography of functional genes shared among seven terrestrial subsurface metagenomes reveal N-cycling and microbial evolutionary relationships. Front Microbiol 5: 531.
Lin LH, Wang PL, Rumble D, Lippmann-Pipke J, Boice E, Pratt LM et al (2006). Long-term sustainability of a high-energy, low-diversity crustal biome. Science 314: 479–482.
Lin X, Kennedy D, Fredrickson J, Bjornstad B, Konopka A (2012). Vertical stratification of subsurface microbial community composition across geological formations at the Hanford Site. Environ Microbiol 14: 414–425.
Lollar BS, Lacrampe-Couloume G, Slater GF, Ward J, Moser DP, Gihring TM et al (2006). Unravelling abiogenic and biogenic sources of methane in the Earth's deep subsurface. Chem Geol 22: 328–339.
Lovley DR, Chapelle FH (1995). Deep subsurface microbial processes. Rev Geophys 33: 365–381.
Loy A, Duller S, Baranyi C, Mußmann M, Ott J, Sharon I et al (2009). Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur‐oxidizing prokaryotes. Environ Microbiol 11: 289–299.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A et al (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
McMahon S, Parnell J (2014). Weighing the deep continental biosphere. FEMS Microbiol Ecol 87: 113–120.
Magnabosco C, Ryan K, Lau MC, Kuloyo O, Lollar BS, Kieft TL et al (2015). A metagenomic window into carbon metabolism at 3 km depth in Precambrian continental crust. ISME J 10: 730–741.
Matsen FA, Kodner RB, Armbrust EV (2010). pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11: 1–16.
Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D et al (2008). IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res 36: 534–538.
Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Pillay M et al (2014). IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res 42: 568–573.
Momper LM, Reese BK, Carvalho G, Lee P, Webb EA (2015). A novel cohabitation between two diazotrophic cyanobacteria in the oligotrophic ocean. ISME J 4: 882–893.
Nyyssönen M, Hultman J, Ahonen L, Kukkonen I, Paulin L, Laine P et al (2014). Taxonomically and functionally diverse microbial communities in deep crystalline rocks of the Fennoscandian shield. ISME J 8: 126–138.
Onstott TC, Phelps TJ, Colwell FS, Ringelberg D, White DC, Boone DR et al (1998). Observations pertaining to the origin and ecology of microorganisms recovered from the deep subsurface of Taylorsville Basin, Virginia. Geomicrobiol J 15: 353–585.
Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev 75: 361–422.
Osburn MR, LaRowe DE, Momper L, Amend JP (2014). Chemolithotrophy in the continental deep subsurface: Sanford Underground Research Facility (SURF), USA. Front Extr Microbiol 5: 610.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Gen Res 25: 1043–1055.
Peng Y, Leung HC, Yiu SM, Chin FY (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428.
Pedersen K (2000). Exploration of deep intraterrestrial microbial life: current perspectives. FEMS Microbiol Lett 185: 9–16.
Pfiffner SM, Cantu JM, Smithgall A, Peacock AD, White DC, Moser DP et al (2006). Deep subsurface microbial biomass and community structure in Witwatersrand Basin mines. Geomicrobiol J 23: 431–442.
Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH et al (2008). Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol 190: 3192–3202.
Price MN, Dehal PS, Arkin AP (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490.
Pruesse E, Queast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Pruesse E, Peplies J, Glöckner FO (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: 590–596.
Rabus R, Hansen TA, Widdel F (2006). Dissimilatory sulfate- and sulfur-reducing prokaryotes. The Prokaryotes Springer: New York, NY, USA, pp 659–768.
Reed AJ, Lutz RA, Vetriani C (2006). Vertical distribution and diversity of bacteria and archaea in sulfide and methane-rich cold seep sediments located at the base of the Florida Escarpment. Extremophiles 10: 199–211.
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF et al (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437.
Sander J, Engels-Schwarzlose S, Dahl C (2006). Importance of the DsrMKJOP complex for sulfur oxidation in Allochromatium vinosum and phylogenetic analysis of related complexes in other prokaryotes. Arch Microbiol 186: 357–366.
Schauder R, Preuß A, Jetten M, Fuchs G (1988). Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum. Arch Microbiol 151: 84–89.
Seitz KW, Lazar CS, Hinrichs K-U, Teske AP, Baker BJ (2016). Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J 10: 1696–1705.
Simkus DN, Slater GF, Lollar BS, Wilkie K, Kieft TL, Magnabosco C et al (2016). Variations in microbial carbon sources and cycling in the deep continental subsurface. Geochimica et Cosmochimica Acta 173: 264–283.
Speth DR, Guerrero-Cruz S, Dutilh BE, Jetten MS (2016). Genome-based microbial ecology of anammox granules in a full-scale wastewater treatment system. Nature Communications 7: 1–10.
Stamatakis A (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Stevens T (1997). Lithoautotrophy in the subsurface. FEMS Microbiol Rev 20: 327–337.
Stevens TO, McKinley JP (1995). Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science 270: 450–454.
Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS (2008). Distinct form I, II, III, and IV RuBisCo proteins from the three kingdoms of life provide clues about RuBisCo evolution and structure/function relationships. J Exp Botany 59: 1515–1524.
Tiago I, Veríssimo A (2013). Microbial and functional diversity of a subterrestrial high pH groundwater associated to serpentinization. Environ Microbiol 15: 1687–1706.
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43.
Wagner M, Horn M (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17: 241–249.
Whitman WB, Coleman DC, Wiebe WJ (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583.
Wu M, Scott AJ (2012). Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28: 1033–1034.
Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH et al (2008). The All-Species Living Tree project: A 16 S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 31: 241–250.
Youssef NH, Rinke C, Stepanauskas R, Farag I, Woyke T, Elshahed MS (2015a). Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum 'Diapherotrites'. ISME J 9: 447–460.
Acknowledgements
This work was supported by the NASA Astrobiology Institute under cooperative agreement NNA13AA92A. Many thanks to John Heidelberg and Rohan Sachdeva at USC for use of their servers and help in metagenomic analysis. We would also like to recognize A Murat Eren for his invaluable help in utilizing the Anvi’o interface. We want to thank especially staff and personnel at SURF for access to the deep subsurface and repeated access to samples used in this study. This work was funded by the NASA Astrobiology Institute under cooperative agreement NNA13AA92A.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Momper, L., Jungbluth, S., Lee, M. et al. Energy and carbon metabolisms in a deep terrestrial subsurface fluid microbial community. ISME J 11, 2319–2333 (2017). https://doi.org/10.1038/ismej.2017.94
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.94
This article is cited by
-
Methane- and hydrogen-dependent prokaryotic deep biosphere at the Suwa Basin, Japan: impacts of hydrogeological processes on subsurface prokaryotic ecology at the boundary between the North American and the Eurasian Plates
Progress in Earth and Planetary Science (2025)
-
Biosignatures of an ancient bedrock- and impact structure-hosted deep biosphere: current knowledge and future perspectives
Discover Geoscience (2025)
-
Genomic representativeness and chimerism in large collections of SAGs and MAGs of marine prokaryoplankton
Microbiome (2024)
-
Candidatus Desulforudis audaxviator dominates a 975 m deep groundwater community in central Sweden
Communications Biology (2024)
-
Metabolic adaptations underpin high productivity rates in relict subsurface water
Scientific Reports (2024)