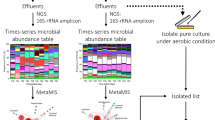
Abstract
Ecosystem development theory predicts that successional turnover in community composition can influence ecosystem functioning. However, tests of this theory in natural systems are made difficult by a lack of replicable and tractable model systems. Using the microbial digestive associates of a carnivorous pitcher plant, I tested hypotheses linking host age-driven microbial community development to host functioning. Monitoring the yearlong development of independent microbial digestive communities in two pitcher plant populations revealed a number of trends in community succession matching theoretical predictions. These included mid-successional peaks in bacterial diversity and metabolic substrate use, predictable and parallel successional trajectories among microbial communities, and convergence giving way to divergence in community composition and carbon substrate use. Bacterial composition, biomass, and diversity positively influenced the rate of prey decomposition, which was in turn positively associated with a host leaf’s nitrogen uptake efficiency. Overall digestive performance was greatest during late summer. These results highlight links between community succession and ecosystem functioning and extend succession theory to host-associated microbial communities.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Agresti A . (2013) Categorical Data Analysis, 3rd ed. John Wiley & Sons, Inc.: Hoboken, NJ, USA.
Alday JG, Marrs RH, Martínez-Ruiz C . (2011). Vegetation convergence during early succession on coal wastes: a 6-year permanent plot study. J Veg Sci 22: 1072–1083.
Anderson MJ . (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
Armitage DW . (2016). Time-variant species pools shape competitive dynamics and biodiversity–ecosystem function relationships. Proc R Soc B 283.
Auclair AN, Goff FG . (1971). Diversity relations of upland forests in the Western Great Lakes area. Am Nat 105: 499–528.
Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI . (2005). Host-bacterial mutualism in the human intestine. Science 307: 1915–1920.
Balser TC, Firestone MK . (2005). Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry 73: 395–415.
Beaver RA . (1985). Geographical variation in food web structure in Nepenthes pitcher plants. Ecol Entomol 10: 241–248.
Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK . (2005). The contribution of species richness and composition to bacterial services. Nature 436: 1157–1160.
Burnham KP, Anderson DR . (2003) Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. Springer Science & Business Media: Berlin, Germany.
Butler JL, Gotelli NJ, Ellison AM . (2008). Linking the brown and green: nutrient transformation and fate in the Sarracenia microecosystem. Ecology 89: 898–904.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Cardinale BJ, Wright JP, Cadotte MW, Carroll IT, Hector A, Srivastava DS et al. (2007). Impacts of plant diversity on biomass production increase through time because of species complementarity. Proc Natl Acad Sci USA 104: 18123–18128.
Christensen NL, Peet RK . (1984). Convergence during secondary forest succession. J Ecol 72: 25–36.
Connell JH, Slatyer RO . (1977). Mechanisms of succession in natural communities and their role in community stability and organization. Am Nat 111: 1119–1144.
Copeland JK, Yuan L, Layeghifard M, Wang PW, Guttman DS . (2015). Seasonal community succession of the phyllosphere microbiome. Mol Plant Microbe Interact 28: 274–285.
DeAngelis DL . (1992) Dynamics of Nutrient Cycling and Food Webs. Chapman & Hall: London, UK; New York, NY, USA.
Dini-Andreote F, Stegen JC, Elsas JD, van, Salles JF . (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci USA 112: E1326–E1332.
Eisenhauer N, Schulz W, Scheu S, Jousset A . (2013). Niche dimensionality links biodiversity and invasibility of microbial communities. Funct Ecol 27: 282–288.
Evans S, Martiny JBH, Allison SD . (2017). Effects of dispersal and selection on stochastic assembly in microbial communities. ISME J 11: 176–185.
Fierer N, Nemergut D, Knight R, Craine JM . (2010). Changes through time: integrating microorganisms into the study of succession. Res Microbiol 161: 635–642.
Finn JT . (1982). Ecosystem succession, nutrient cycling and output-input ratios. J Theor Biol 99: 479–489.
Fisher SG, Gray LJ, Grimm NB, Busch DE . (1982). Temporal succession in a desert stream ecosystem following flash flooding. Ecol Monogr 52: 93–110.
Graham EB, Knelman JE, Schindlbacher A, Siciliano S, Breulmann M, Yannarell A et al. (2016). Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front Microbiol 7: 214.
Harmon LJ, Matthews B, Des Roches S, Chase JM, Shurin JB, Schluter D . (2009). Evolutionary diversification in stickleback affects ecosystem functioning. Nature 458: 1167–1170.
Hättenschwiler S, Fromin N, Barantal S . (2011). Functional diversity of terrestrial microbial decomposers and their substrates. C R Biol 334: 393–402.
Hooper DU, Chapin FS, Ewel JJ, Hector A, Inchausti P, Lavorel S et al. (2005). Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol Monogr 75: 3–35.
Huston M, Smith T . (1987). Plant succession: life history and competition. Am Nat 130: 168–198.
Jemielita M, Taormina MJ, Burns AR, Hampton JS, Rolig AS, Guillemin K et al. (2014). Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio 5: e01751–14.
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M . (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44: D457–D462.
Klappenbach JA, Dunbar JM, Schmidt TM . (2000). rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol 66: 1328–1333.
Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 108: 4578–4585.
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16 S rRNA marker gene sequences. Nat Biotechnol 31: 814–821.
Livermore JA, Emrich SJ, Tan J, Jones SE . (2014). Freshwater bacterial lifestyles inferred from comparative genomics. Environ Microbiol 16: 746–758.
Loreau M . (1998). Ecosystem development explained by competition within and between material cycles. Proc R Soc B Biol Sci 265: 33–38.
Loreau M . (2001). Microbial diversity, producer–decomposer interactions and ecosystem processes: a theoretical model. Proc R Soc Lond B Biol Sci 268: 303–309.
Loucks OL . (1970). Evolution of diversity, efficiency, and community stability. Am Zool 10: 17–25.
Love MI, Huber W, Anders S . (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 1–21.
Lugtenberg B, Kamilova F . (2009). Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63: 541–556.
Marino S, Baxter NT, Huffnagle GB, Petrosino JF, Schloss PD . (2014). Mathematical modeling of primary succession of murine intestinal microbiota. Proc Natl Acad Sci USA 111: 439–444.
McMurdie PJ, Holmes S . (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE 8: e61217.
Meiners SJ, Pickett STA, Cadenasso ML . (2015) An Integrative Approach to Successional Dynamics: Tempo and Mode of Vegetation Change. Cambridge University Press: Cambridge, UK.
Metcalf JL, Xu ZZ, Weiss S, Lax S, Treuren WV, Hyde ER et al. (2016). Microbial community assembly and metabolic function during mammalian corpse decomposition. Science 351: 158–162.
Miller TE, terHorst CP . (2012). Testing successional hypotheses of stability, heterogeneity, and diversity in pitcher-plant inquiline communities. Oecologia 170: 243–251.
Moorhead DL, Hall DL, Willig MR . (1998). Succession of macroinvertebrates in playas of the Southern High Plains, USA. J North Am Benthol Soc 17: 430–442.
Nemergut DR, Knelman JE, Ferrenberg S, Bilinski T, Melbourne B, Jiang L et al. (2015). Decreases in average bacterial community rRNA operon copy number during succession. ISME J 10: 1147–1156.
Neutel A-M, Heesterbeek JAP, van de Koppel J, Hoenderboom G, Vos A, Kaldeway C et al. (2007). Reconciling complexity with stability in naturally assembling food webs. Nature 449: 599–602.
Nielsen UN, Ayres E, Wall DH, Bardgett RD . (2011). Soil biodiversity and carbon cycling: a review and synthesis of studies examining diversity–function relationships. Eur J Soil Sci 62: 105–116.
Odum EP . (1969). The strategy of ecosystem development. Science 164: 262–270.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB et al. (2015), vegan: Community Ecology Package.
Orrock JL, Watling JI . (2010). Local community size mediates ecological drift and competition in metacommunities. Proc R Soc Lond B Biol Sci 277: 2185–2191.
Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO . (2007). Development of the human infant intestinal microbiota. PLOS Biol 5: e177.
Peet RK, Christensen NL . (1988) Changes in species diversity during secondary forest succession on the North Carolina Piedmont. In: During HJ, Werger MJA, Williams JH (eds). Diversity and Pattern in Plant Communities. SPB Academic Publishing: The Hague, The Netherlands, pp 233–245.
R Development Core Team. (2015) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Redford AJ, Fierer N . (2009). Bacterial succession on the leaf surface: a novel system for studying successional dynamics. Microb Ecol 58: 189–198.
van Ruijven J, Berendse F . (2005). Diversity-productivity relationships: initial effects, long-term patterns, and underlying mechanisms. Proc Natl Acad Sci USA 102: 695–700.
Schmidt SK, Costello EK, Nemergut DR, Cleveland CC, Reed SC, Weintraub MN et al. (2007). Biogeochemical consequences of rapid microbial turnover and seasonal succession in soil. Ecology 88: 1379–1385.
Strickland MS, Lauber C, Fierer N, Bradford MA . (2009). Testing the functional significance of microbial community composition. Ecology 90: 441–451.
Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611.
Thompson IP, Bailey MJ, Fenlon JS, Fermor TR, Lilley AK, Lynch JM et al. (1993). Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris). Plant Soil 150: 177–191.
Tilman D . (1990). Constraints and tradeoffs: toward a predictive theory of competition and succession. Oikos 58: 3–15.
Vellend M . (2016) The Theory of Ecological Communities. Princeton University Press: Princeton, NJ, USA.
Vitousek PM, Reiners WA . (1975). Ecosystem succession and nutrient retention: a hypothesis. Bioscience 25: 376–381.
Weis JJ, Cardinale BJ, Forshay KJ, Ives AR . (2007). Effects of species diversity on community biomass production change over the course of succession. Ecology 88: 929–939.
Williams MA, Jangid K, Shanmugam SG, Whitman WB . (2013). Bacterial communities in soil mimic patterns of vegetative succession and ecosystem climax but are resilient to change between seasons. Soil Biol Biochem 57: 749–757.
Wolfe LM . (1981). Feeding behavior of a plant: differential prey capture in old and new leaves of the pitcher plant (Sarracenia purpurea. Am Midl Nat 106: 352–359.
Zhou J, Deng Y, Zhang P, Xue K, Liang Y, Nostrand JDV et al. (2014). Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci USA 111: E836–E845.
Acknowledgements
I thank Anna Petrosky, Ramon Leon, & Stefani Brandt for assistance with data collection. Ellen Simms, Todd Dawson, and the UC Berkeley Forestry Camp provided facilities and equipment. I thank Stuart Jones, Mary Firestone, Mary Power & Wayne Sousa for critical feedback. Field collection permits were provided by Jim Belsher-Howe (USFS). Data are publically available on the MG-RAST server under project ID mgp14344. Funding was provided by NSF DEB-1406524 & an NSF GRFP.
Author contributions
DWA conceived this work, performed data collection and analysis, and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Armitage, D. Linking the development and functioning of a carnivorous pitcher plant’s microbial digestive community. ISME J 11, 2439–2451 (2017). https://doi.org/10.1038/ismej.2017.99
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.99
This article is cited by
-
An acidophilic fungus promotes prey digestion in a carnivorous plant
Nature Microbiology (2024)
-
Bacterial communities in carnivorous pitcher plants colonize and persist in inquiline mosquitoes
Animal Microbiome (2022)
-
Tropical pitcher plants (Nepenthes) act as ecological filters by altering properties of their fluid microenvironments
Scientific Reports (2020)
-
Context-dependent dynamics lead to the assembly of functionally distinct microbial communities
Nature Communications (2020)
-
Characterizing differences in microbial community composition and function between Fusarium wilt diseased and healthy soils under watermelon cultivation
Plant and Soil (2019)