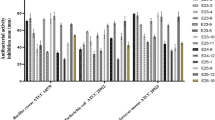
Abstract
The bacterial genus Streptomyces is endowed with a remarkable secondary metabolism that generates an enormous number of bioactive small molecules. Many of these genetically encoded small molecules are used as antibiotics, anticancer agents and as other clinically relevant therapeutics. The rise of resistant pathogens has led to calls for renewed efforts to identify antimicrobial activities, including expanded screening of streptomycetes. Indeed, it is known that most strains encode >20 secondary metabolites and that many, perhaps most of these, have not been considered for their possible therapeutic use. One roadblock is that many strains do not express their secondary metabolic gene clusters efficiently under laboratory conditions. As one approach to this problem, we have used alleles of a pleiotropic regulator of secondary metabolism from Streptomyces coelicolor to activate secondary biosynthetic gene clusters in heterologous streptomycetes. In one case, we demonstrate the activation of pulvomycin production in S. flavopersicus, a metabolite not previously attributed to this species. We find that the absA1-engineered strains produced sufficient material for purification and characterization. As a result, we identified new, broad-spectrum antimicrobial activities for pulvomycin, including a potent antimicrobial activity against highly antibiotic-resistant Gram-negative and Gram-positive pathogens.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Baltz, R. H. Antimicrobials from Actinomycetes: Back to the Future. Microbe. 2, 6–7 (2007).
Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002).
Ikeda, H. et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21, 526–531 (2003).
Ohnishi, Y. et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060 (2008).
Adamidis, T., Riggle, P. & Champness, W. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J. Bacteriol. 172, 2962–2969 (1990).
Brian, P., Riggle, P. J., Santos, R. A. & Champness, W. C. Global negative regulation of Streptomyces coelicolor antibiotic synthesis mediated by an absA-encoded putative signal transduction system. J. Bacteriol. 178, 3221–3231 (1996).
Sheeler, N. L., MacMillan, S. V. & Nodwell, J. R. Biochemical activities of the absA two-component system of Streptomyces coelicolor. J. Bacteriol. 187, 687–696 (2005).
McKenzie, N. L. & Nodwell, J. R. Transmembrane topology of the AbsA1 sensor kinase of Streptomyces coelicolor. Microbiology 155, 1812–1818 (2009).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces genetics (The John Innes Foundation, Norwich, UK, 2000).
Guglierame, P. et al. Efflux pump genes of the resistance-nodulation-division family in Burkholderia cenocepacia genome. BMC Microbiol. 6, 66 (2006).
Poole, K. & Srikumar, R. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top Med. Chem. 1, 59–71 (2001).
Volff, J. N. & Altenbuchner, J. Genetic instability of the Streptomyces chromosome. Mol. Microbiol. 27, 239–246 (1998).
Cone, M. C., Petrich, A. K., Gould, S. J. & Zabriskie, T. M. Cloning and heterologous expression of blasticidin S biosynthetic genes from Streptomyces griseochromogenes. J. Antibiot (Tokyo) 51, 570–578 (1998).
Mason, D. J., Dietz, A. & Smith, R. M. Actinospectacin, a new antibiotic. I. Discovery and biological properties. Antibiot. Chemother. 11, 118–122 (1961).
Smith, R. Structure revision of the antibiotic pulvomycin. J Am. Chem. Soc. 107, 2849–2859 (1985).
Wolf, H., Assmann, D. & Fischer, E. Pulvomycin, an inhibitor of protein biosynthesis preventing ternary complex formation between elongation factor Tu, GTP, and aminoacyl-tRNA. Proc. Natl Acad. Sci. USA 75, 5324–5328 (1978).
Selva, E. et al. Antibiotic GE2270 a: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization. J. Antibiot. (Tokyo) 44, 693–701 (1991).
Chater, K. F. & Horinouchi, S. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48, 9–15 (2003).
van Wezel, G. P., McKenzie, N. L. & Nodwell, J. R. Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol. 458, 117–141 (2009).
Rigali, S. et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9, 670–675 (2008).
Hosaka, T. et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 27, 462–464 (2009).
Acknowledgements
We thank Susan Jensen and Marie Elliot for helpful comments on this paper. We also thank Mervyn Bibb for communicating unpublished information with us. Work in JRN's laboratory was supported by the Natural Science and Engineering Research Council (165846) and by JNE Biotech Inc. (Hamilton, Ontario). NLM was supported by a Canadian Institutes of Health Research Canada Graduate Scholarship (CGD—80406). Work in GDW's laboratory was supported by the Canadian Institutes for Health Research (MT-14981) and the Natural Science and Engineering Research Council.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
McKenzie, N., Thaker, M., Koteva, K. et al. Induction of antimicrobial activities in heterologous streptomycetes using alleles of the Streptomyces coelicolor gene absA1. J Antibiot 63, 177–182 (2010). https://doi.org/10.1038/ja.2010.13
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ja.2010.13
Keywords
This article is cited by
-
Identification of pulvomycin as an inhibitor of the futalosine pathway
The Journal of Antibiotics (2021)
-
Production of pikromycin using branched chain amino acid catabolism in Streptomyces venezuelae ATCC 15439
Journal of Industrial Microbiology and Biotechnology (2018)
-
Identification of butenolide regulatory system controlling secondary metabolism in Streptomyces albus J1074
Scientific Reports (2017)
-
Competition and co-regulation of spirotoamide and tautomycetin biosynthesis in Streptomyces griseochromogenes, and isolation and structural elucidation of spirotoamide C and D
The Journal of Antibiotics (2017)
-
Metabolic perturbation to enhance polyketide and nonribosomal peptide antibiotic production using triclosan and ribosome-targeting drugs
Applied Microbiology and Biotechnology (2017)