Abstract
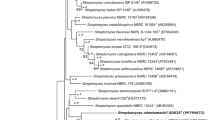
A novel Gram-stain-positive actinobacterium, designated RI148-Li105T, was isolated from a lichen sample from Rishiri Island, Japan, and its taxonomic position was investigated by a polyphasic approach. 16S rRNA gene sequencing study indicated that strain RI148-Li105T was related to the type strain of Luteimicrobium subarcticum, with a similarity of 97.8%. Cells of strain RI148-Li105T exhibited a rod–coccus cycle. The diagnostic cell-wall diamino acid of this organism was lysine and the peptidoglycan type was found to be A4α. The predominant menaquinones were MK-8(H2) and MK-9(H2), and the major fatty acids were iso-C16:0, C17:1 ω9c and C17:0. The DNA G+C content was 73.6 mol%. The major phenotypic characteristics of strain RI148-Li105T basically corresponded to those of the genus Luteimicrobium excluding the fatty acid composition. These results suggest that strain RI148-Li105T should be affiliated with the genus Luteimicrobium. Meanwhile, DNA–DNA hybridization and some phenotypic characteristics revealed that the strain differs from L. subarcticum. Therefore, strain RI148-Li105T represents a novel species of the genus Luteimicrobium, for which the name Luteimicrobium album sp. nov. is proposed. The type strain of Luteimicrobium album is RI148-Li105T (=NBRC 106348T=DSM 24866T).
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Hamada, M. et al. Luteimicrobium subarcticum gen. nov., sp. nov., a new member of the suborder Micrococcineae. Int. J. Syst. Evol. Microbiol. 60, 796–800 (2010).
Schleifer, K. H. & Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477 (1972).
Yamamura, H. et al. Actinomycetospora iriomotensis sp. nov., a novel actinomycete isolated from a lichen sample. J. Antibiot. 64, 289–292 (2011).
Yamamura, H. et al. Actinomycetospora rishiriensis sp. nov., an actinomycete isolated from a lichen. Int. J. Syst. Evol. Microbiol. 61, 2621–2625 (2011).
Cardinale, M., Grube, M. & Berg, G. Frondihabitans cladoniiphilus sp. nov., a novel actinobacterium of the family Microbacteriaceae isolated from lichen, and emended description of the genus Frondihabitans. Int. J. Syst. Evol. Microbiol. 61, 3033–3038 (2011).
An, S. Y., Xiao, T. & Yokota, A. Leifsonia lichenia sp. nov., isolated from lichen in Japan. J. Gen. Appl. Microbiol. 55, 339–343 (2009).
Li, B., Xie, C. H. & Yokota, A. Nocardioides exalbidus sp. nov., a novel actinomycete isolated from lichen in Izu-Oshima Island, Japan. Actinomycetologica 21, 22–26 (2007).
Hayakawa, M. & Nonomura, H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509 (1987).
Gerhardt, P., Murray, R. G. E., Wood, W. A. & Krieg, N. R. Methods for General and Molecular Bacteriology (Edited by American Society for Microbiology) (Washington, DC, 1994).
Chun, J. et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261 (2007).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 738–791 (1985).
Saito, H. & Miura, K. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta. 72, 619–629 (1963).
Tamaoka, J. & Komagata, K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS. Microbiol. Lett. 25, 125–128 (1984).
Ezaki, T. et al. Simple genetic identification method of viridans group streptococci by colorimetric dot hybridization and quantitative fluorometric hybridization in microdilution wells. J. Clin. Microbiol. 26, 1708–1713 (1988).
Ezaki, T., Hashimoto, Y. & Yabuuchi, E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229 (1989).
Tamura, T., Nakagawa, Y. & Kawasaki, H. DNA–DNA hybridization method In Identification Manual of Actinomycetes (Edited by The Society for Actinomycetes Japan) 134–137 (The Business Center for Academic Societies in Japanese, Tokyo, 2001).
Nozawa, Y., Sakai, N., Arai, K., Kawasaki, Y. & Harada, K. Reliable and sensitive analysis of amino acids in the peptidoglycan of actinomycetes using the advanced Marfey’s method. J. Microbiol. Methods 70, 306–311 (2007).
Honda, S. et al. High-performance liquid chromatography of reducing carbohydrates as strongly ultraviolet-absorbing and electrochemically sensitive 1-phenyl-3-methyl-5-pyrazolone derivatives. Anal. Biochem. 180, 351–357 (1989).
Yang, X., Zhao, Y., Wang, Q., Wang, H. & Mei, Q. Analysis of the monosaccharide components in Angelica polysaccharides by high performance liquid chromatography. Anal. Sci. 21, 1177–1180 (2005).
Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids, MIDI Technical Note 101. MIDI Inc. Newark, Delaware (1990).
MIDI MIS operating manual, version 4.5. MIDI, Inc. Newark, Delaware (2002).
Hamada, M. et al. Mobilicoccus pelagius gen. nov., sp. nov. and Piscicoccus intestinalis gen. nov., sp. nov., two new members of the family Dermatophilaceae, and reclassification of Dermatophilus chelonae (Masters et al. 1995) as Austwickia chelonae gen. nov., comb. nov. J. Gen. Appl. Microbiol. 56, 427–436 (2010).
Wayne, L. G. et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Acknowledgements
We are grateful to Mr Yuya Sakuraki for excellent technical assistance. This study was partly supported by a research grant from the Institute for Fermentation, Osaka (IFO), Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Hamada, M., Yamamura, H., Komukai, C. et al. Luteimicrobium album sp. nov., a novel actinobacterium isolated from a lichen collected in Japan, and emended description of the genus Luteimicrobium. J Antibiot 65, 427–431 (2012). https://doi.org/10.1038/ja.2012.45
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ja.2012.45
Keywords
This article is cited by
-
Streptomyces pacificus sp. nov., a novel spongiicolazolicin-producing actinomycete isolated from a coastal sediment
The Journal of Antibiotics (2023)
-
Evaluation of the Potential of Rhizobacteria in Supplying Nutrients of Zea mays L. Plant with a Focus on Zinc
Journal of Soil Science and Plant Nutrition (2023)
-
Effusibacillus dendaii sp. nov. isolated from farm soil
Archives of Microbiology (2021)
-
Chitinophaga agri sp. nov., a bacterium isolated from soil of reclaimed land
Archives of Microbiology (2021)
-
Xylophilus rhododendri sp. nov., Isolated from Flower of Royal Azalea, Rhododendron schlippenbachii
Current Microbiology (2020)