Abstract
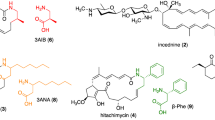
A biosynthetic gene cluster for the 24-membered macrolactam antibiotic incednine was identified from the producer strain, Streptomyces sp. ML694-90F3. Among the putative incednine biosynthetic enzymes, a novel pyridoxal 5′-phosphate (PLP)-dependent β-glutamate-β-decarboxylase, IdnL3, was functionally characterized in vitro by demonstrating its (S)-3-aminobutyrate-forming activity with β-glutamate in the presence of PLP. Because (S)-3-aminobutyrate is known for the direct precursor of incednine, this enzyme supplies the unique β-amino acid starter unit. The identified gene cluster encodes five characteristic β-amino acid carrying enzymes, consisting of a pathway-specific ATP-dependent ligase, a discrete acyl carrier protein (ACP), β-aminoacyl-ACP β-amino group-protecting ATP-dependent ligase, dipeptidyl-ACP:PKS-loading ACP dipeptidyltransferase and a terminal amino acid peptidase, which are completely conserved in β-amino acid-containing macrolactam biosynthetic gene clusters. Overall, a plausible biosynthetic pathway for incednine was proposed.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Futamura, Y. et al. Discovery of incednine as a potent modulator of the anti-apoptotic function of Bcl-xL from microbial origin. J. Am. Chem. Soc 130, 1822–1823 (2008).
Schulz, D. et al. Silvalactam, a 24-membered macrolactam antibiotic produced by Streptomyces sp. Tu 6392. J. Antibiot. 65, 369–372 (2012).
Takaishi, M., Kudo, F. & Eguchi, T. Biosynthetic pathway of 24-membered macrolactam glycoside incednine. Tetrahedron 64, 6651–6656 (2008).
Takaishi, M., Kudo, F. & Eguchi, T. A unique pathway for the 3-aminobutyrate starter unit from L-glutamate through b-glutamate during biosynthesis of the 24-membered macrolactam antibiotic, incednine. Org. Lett. 14, 4591–4593 (2012).
Ruzicka, F. J. & Frey, P. A. Glutamate 2,3-aminomutase: a new member of the radical SAM superfamily of enzymes. Biochim. Biophys. Acta 1774, 286–296 (2007).
Ogasawara, Y., Kakinuma, K. & Eguchi, T. Involvement of glutamate mutase in the biosynthesis of the unique starter unit of the macrolactam polyketide antibiotic vicenistatin. J. Antibiot. 58, 468–472 (2005).
Udwary, D. W. et al. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA. 104, 10376–10381 (2007).
Jorgensen, H. et al. Biosynthesis of macrolactam BE-14106 involves two distinct PKS systems and amino acid processing enzymes for generation of the aminoacyl starter unit. Chem. Biol. 16, 1109–1121 (2009).
Jorgensen, H. et al. Insights into the evolution of macrolactam biosynthesis through cloning and comparative analysis of the biosynthetic gene cluster for a novel macrocyclic lactam, ML-449. Appl. Environ. Microbiol. 76, 283–293 (2010).
Shinohara, Y., Kudo, F. & Eguchi, T. A natural protecting group strategy to carry an amino acid starter unit in the biosynthesis of macrolactam polyketide antibiotics. J. Am. Chem. Soc. 133, 18134–18137 (2011).
Kudo, F., Kitayama, T., Kakinuma, K. & Eguchi, T. Macrolactam formation catalyzed by the thioesterase domain of vicenistatin polyketide synthase. Tetrahedron Lett. 47, 1529–1532 (2006).
Carroll, B. J. et al. Identification of a set of genes involved in the formation of the substrate for the incorporation of the unusual ‘glycolate’ chain extension unit in ansamitocin biosynthesis. J. Am. Chem. Soc. 124, 4176–4177 (2002).
Haydock, S. F. et al. Organization of the biosynthetic gene cluster for the macrolide concanamycin A in Streptomyces neyagawaensis ATCC 27449. Microbiology 151, 3161–3169 (2005).
Zhao, C. et al. Utilization of the methoxymalonyl-acyl carrier protein biosynthesis locus for cloning the oxazolomycin biosynthetic gene cluster from Streptomyces albus JA3453. J. Bacteril. 188, 4142–4147 (2006).
Karki, S. et al. The methoxymalonyl-acyl carrier protein biosynthesis locus and the nearby gene with the b-ketoacyl synthase domain are involved in the biosynthesis of galbonolides in Streptomyces galbus, but these loci are separate from the modular polyketide synthase gene cluster. FEMS Microbiol. Lett. 310, 69–75 (2010).
Chan, Y. A., Podevels, A. M., Kevany, B. M. & Thomas, M. G. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 26, 90–114 (2009).
Haydock, S. F. et al. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 374, 246–248 (1995).
Zhang, F. et al. Cloning and elucidation of the FR901464 gene cluster revealing a complex acyltransferase-less polyketide synthase using glycerate as starter units. J. Am. Chem. Soc. 133, 2452–2462 (2011).
Nedal, A. & Zotchev, S. B. Biosynthesis of deoxyaminosugars in antibiotic-producing bacteria. Appl. Microbiol. Biotechnol. 64, 7–15 (2004).
Trefzer, A., Salas, J. A. & Bechthold, A. Genes and enzymes involved in deoxysugar biosynthesis in bacteria. Nat. Prod. Rep. 16, 283–299 (1999).
Hong, L., Zhao, Z., Melançon, C. E. III, Zhang, H. & Liu, H. W. In vitro characterization of the enzymes involved in TDP-D-forosamine biosynthesis in the spinosyn pathway of Saccharopolyspora spinosa. J. Am. Chem. Soc. 130, 4954–4967 (2008).
Yu, T. W. et al. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. USA. 99, 7968–7973 (2002).
Lohman, J. R. et al. Cloning and sequencing of the kedarcidin biosynthetic gene cluster from Streptoalloteichus sp. ATCC 53650 revealing new insights into biosynthesis of the enediyne family of antitumor antibiotics. Mol. Biosyst. 9, 478–491 (2013).
Ansari, M. Z., Yadav, G., Gokhale, R. S. & Mohanty, D. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic. Acids. Res. 32, W405–W413 (2004).
Tang, L., Yoon, Y. J., Choi, C. Y. & Hutchinson, C. R. Characterization of the enzymatic domains in the modular polyketide synthase involved in rifamycin B biosynthesis by Amycolatopsis mediterranei. Gene 216, 255–265 (1998).
Del Vecchio, F. et al. Active-site residue, domain and module swaps in modular polyketide synthases. J. Ind. Microbiol. Biotechnol. 30, 489–494 (2003).
Keatinge-Clay, A. T. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol. 14, 898–908 (2007).
Keatinge-Clay, A. Crystal structure of the erythromycin polyketide synthase dehydratase. J. Mol. Biol. 384, 941–953 (2008).
Keatinge-Clay, A. T. & Stroud, R. M. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure 14, 737–748 (2006).
Caffrey, P. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. Chembiochem 4, 654–657 (2003).
Walton, L. J., Corre, C. & Challis, G. L. Mechanisms for incorporation of glycerol-derived precursors into polyketide metabolites. J. Ind. Microbiol. Biotechnol. 33, 105–120 (2006).
Dorrestein, P. C. et al. The bifunctional glyceryl transferase/phosphatase OzmB belonging to the HAD superfamily that diverts 1,3-bisphosphoglycerate into polyketide biosynthesis. J. Am. Chem. Soc. 128, 10386–10387 (2006).
Watanabe, K., Khosla, C., Stroud, R. M. & Tsai, S. C. Crystal structure of an Acyl-ACP dehydrogenase from the FK520 polyketide biosynthetic pathway: insights into extender unit biosynthesis. J. Mol. Biol. 334, 435–444 (2003).
Waldron, C. et al. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem. Biol. 8, 487–499 (2001).
Hong, L., Zhao, Z. & Liu, H. W. Characterization of SpnQ from the spinosyn biosynthetic pathway of Saccharopolyspora spinosa: mechanistic and evolutionary implications for C-3 deoxygenation in deoxysugar biosynthesis. J. Am. Chem. Soc. 128, 14262–14263 (2006).
Zhao, Z., Hong, L. & Liu, H. W. Characterization of protein encoded by spnR from the spinosyn gene cluster of Saccharopolyspora spinosa: mechanistic implications for forosamine biosynthesis. J. Am. Chem. Soc. 127, 7692–7693 (2005).
Gu, X., Glushka, J., Lee, S. G. & Bar-Peled, M. Biosynthesis of a new UDP-sugar, UDP-2-acetamido-2-deoxyxylose, in the human pathogen Bacillus cereus subspecies cytotoxis NVH 391-98. J. Biol. Chem. 285, 24825–24833 (2010).
Claesson, M., Siitonen, V., Dobritzsch, D., Metsa-Ketela, M. & Schneider, G. Crystal structure of the glycosyltransferase SnogD from the biosynthetic pathway of nogalamycin in Streptomyces nogalater. FEBS J. 279, 3251–3263 (2012).
Isiorho, E. A., Liu, H. W. & Keatinge-Clay, A. T. Structural studies of the spinosyn rhamnosyltransferase, SpnG. Biochemistry 51, 1213–1222 (2012).
Munro, A. W., Girvan, H. M. & McLean, K. J. Variations on a (t)heme–novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat. Prod. Rep. 24, 585–609 (2007).
Denisov, I. G., Makris, T. M., Sligar, S. G. & Schlichting, I. Structure and chemistry of cytochrome P 450. Chem. Rev. 105, 2253–2277 (2005).
Borisova, S. A. & Liu, H. W. Characterization of glycosyltransferase DesVII and its auxiliary partner protein DesVIII in the methymycin/picromycin biosynthetic pathway. Biochemistry 49, 8071–8084 (2010).
Leimkuhler, C. et al. Characterization of rhodosaminyl transfer by the AknS/AknT glycosylation complex and its use in reconstituting the biosynthetic pathway of aclacinomycin A. J. Am. Chem. Soc. 129, 10546–10550 (2007).
Baltz, R. H. Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J. Ind. Microbiol. Biotechnol. 38, 1747–1760 (2011).
Zhang, W., Heemstra, J. R. Jr, Walsh, C. T. & Imker, H. J. Activation of the pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry 49, 9946–9947 (2010).
Heathcote, M. L., Staunton, J. & Leadlay, P. F. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 8, 207–220 (2001).
Koglin, A. et al. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature 454, 907–911 (2008).
Claxton, H. B., Akey, D. L., Silver, M. K., Admiraal, S. J. & Smith, J. L. Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway. J. Biol. Chem. 284, 5021–5029 (2009).
Hur, Y. A., Choi, S. S., Sherman, D. H. & Kim, E. S. Identification of TmcN as a pathway-specific positive regulator of tautomycetin biosynthesis in Streptomyces sp. CK4412. Microbiology 154, 2912–2919 (2008).
Laureti, L. et al. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA. 108, 6258–6263 (2011).
Ramos, J. L. et al. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69, 326–356 (2005).
Sletta, H. et al. Nystatin biosynthesis and transport: nysH and nysG genes encoding a putative ABC transporter system in Streptomyces noursei ATCC 11455 are required for efficient conversion of 10-deoxynystatin to nystatin. Antimicrob. Agents Chemother. 49, 4576–4583 (2005).
Toney, M. D. Controlling reaction specificity in pyridoxal phosphate enzymes. Biochim. Biophys. Acta 1814, 1407–1418 (2011).
Lima, S. et al. The crystal structure of the Pseudomonas dacunhae aspartate-b-decarboxylase dodecamer reveals an unknown oligomeric assembly for a pyridoxal-5'-phosphate-dependent enzyme. J. Mol. Biol. 388, 98–108 (2009).
Acknowledgements
We thank Drs Sawa, Igarashi, Takahashi and Akamatsu at the Microbial Chemistry Research Center for providing us with the incednine producer Streptomyces sp. ML694-90F3, and Profs M Kobayashi (The University of Tsukuba, Japan) and K Arakawa (Hiroshima University, Japan) for providing plasmid pHSA81. This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology, the Nagase Science and Technology Foundation, and the Takeda Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Takaishi, M., Kudo, F. & Eguchi, T. Identification of the incednine biosynthetic gene cluster: characterization of novel β-glutamate-β-decarboxylase IdnL3. J Antibiot 66, 691–699 (2013). https://doi.org/10.1038/ja.2013.76
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ja.2013.76
Keywords
This article is cited by
-
Biosynthetic pathway of peucemycin and identification of its derivative from Streptomyces peucetius
Applied Microbiology and Biotechnology (2023)
-
Biosynthetic engineering of the antifungal, anti-MRSA auroramycin
Microbial Cell Factories (2020)
-
Generation of incednine derivatives by mutasynthesis
The Journal of Antibiotics (2020)
-
The many roles of glutamate in metabolism
Journal of Industrial Microbiology and Biotechnology (2016)