Abstract
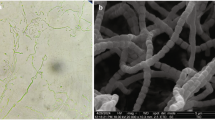
A polyphasic approach was used to determine the taxonomic position of actinomycete strain R1-NS-10T, which was isolated from a sample of strawberry root rhizosphere obtained from Hokuto, Yamanashi, Japan. Strain R1-NS-10T was Gram-staining-positive and aerobic, and formed brownish-white aerial mycelia and grayish-brown substrate mycelia on ISP-2 medium. The strain grew in the presence of 0–5% (w/v) NaCl and optimally grew without NaCl. The strain grew at pH 5–8, and the optimum for growth was pH 7. The optimal growth temperature was 30 °C, but the strain grew at 5–37 °C. Whole-cell hydrolysates of strain R1-NS-10T contained A2pm, galactose, mannose and rhamnose. The predominant menaquinones were MK-9(H6) and MK-9(H8). The major cellular fatty acids were anteiso-C15:0 and iso-C16:0. Comparative 16S rRNA gene sequence analysis revealed that strain R1-NS-10T was most closely related to Streptomyces prunicolor NBRC 13075T (99.4%). The draft genome sequences of both strains were determined for characterization of genome sequence-related parameters such as average nucleotide identity (ANI) and the diversity of secondary metabolite biosynthetic gene clusters. DNA–DNA hybridization (DDH) and ANI values for both strains were below the species delineation cutoff, and differences in physiological and biochemical characteristics differentiated strain R1-NS-10T from its closest phylogenetic relative. On the basis of these data, we propose that strain R1-NS-10T (=NBRC 108812T=KCTC 29186T) should be classified as the type strain of a novel Streptomyces species named Streptomyces hokutonensis sp. nov.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Kämpfer, P. & Genus, I. in Bergey’s Manual of Systematic Bacteriology 2nd edn Vol. 3 eds Goodfellow M., et al1455–1767 Springer: USA, (2012).
Euzéby, J. P. List of Prokaryotic names with Standing in Nomenclature http://www.bacterio.cict.fr/ (2013).
Bérdy, J. Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. 65, 385–395 (2012).
Kino, T. et al. FK-506, a novel immunosuppressant isolated from a Streptomyces. I. Fermentation, isolation, and physico-chemical characteristics. J. Antibiot. 40, 1249–1255 (1987).
Burg, R. W. et al. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15, 361–367 (1979).
Hata, T. et al. Mitomycin, a new antibiotic from Streptomyces. I. J. Antibiot. 9, 141–146 (1956).
Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002).
Ikeda, H. et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21, 526–531 (2003).
Ohnishi, Y. et al. Genome sequence of the streptomycin-producing a microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060 (2008).
Goris, J. et al. DNA-DNA hybridization values and their relationship to whole- genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91 (2007).
Richter, M. & Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl Acad. Sci. USA 106, 19126–19131 (2009).
Konstantinidis, K. T. & Tiedje, J. M. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 10, 504–509 (2007).
Kämpfer, P. in The family Streptomycetaceae. Part I. Taxonomy. in The Prokaryotes: A Handbook on the Biology of Bacteria 3rd edn Vol. 3 eds Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E., 538–604 Springer: New York, (2006).
González, I., Ayuso-Sacido, A., Anderson, A. & Genilloud, O. Actinomycetes isolated from lichens: evaluation of their diversity and detection of biosynthetic gene sequences. FEMS Microbiol. Ecol. 54, 401–415 (2005).
De Vasconcellos, R. L. F. & Cardoso, E. J. B. N. Rhizospheric streptomycetes as potential biocontrol agents of Fusarium and Armillaria pine rot and as PGPR for Pinus taeda. BioControl 54, 807–816 (2009).
Sadeghi, A. et al. Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World J. Microbiol. Biotechnol. 28, 1503–1509 (2012).
Chun, J. et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261 (2007).
Kämpfer, P., Huber, B., Buczolits, S., Thummes, K., Grün-Wollny, I. & Busse, H. J. Streptomyces specialis sp. nov. Int. J. Syst. Evol. Microbiol. 58, 2602–2606 (2008).
Guo, Y., Zheng, W., Rong, X. & Huang, Y. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 58, 149–159 (2008).
Labeda, D. P. Multilocus sequence analysis of phytopathogenic species of the genus Streptomyces. Int. J. Syst. Evol. Microbiol. 61, 2525–2531 (2011).
Rong, X. & Huang, Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst. Appl. Microbiol. 35, 7–18 (2012).
Wayne, L. G. et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Hayakawa, M. & Nonomura, H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509 (1987).
Hayakawa, M., Iino, S. & Nonomura, H. Heavy metal resistance and melanoid pigment production in the streptomycete flora of copper-polluted vineyard soils. J. Ferment. Technol. 60, 1–9 (1982).
Nonomura, H., Iino, S. & Hayakawa, M. Classification of actinomycete genus Ampullariella from soils of Japan. J. Ferment. Technol. 57, 79–85 (1979).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Gerhardt, P. Manual of Methods for General Bacteriology, American Society for Microbiology: Washington, DC, (1981).
Williams, S. T. et al. Numerical classification of Streptomyces and related taxa. J. Gen. Microbiol. 129, 1743–1813 (1983).
Matsukawa, E., Nakagawa, Y., Iimura, Y. & Hayakawa, M. Stimulatory effect of indole-3-acetic acid on aerial mycelium formation and antibiotic production in Streptomyces spp. Actinomycetol. 21, 32–39 (2007).
Gordon, S. A. & Weber, R. P. Colorimetric estimation of indoleacetic acid. Plant. Physiol. 26, 192–195 (1951).
Gordon, R. E. & Mihm, J. M. Identification of Nocardia caviae (Erikson) nov. comb. Ann. N. Y. Acad. Sci. 98, 628–636 (1962).
Hasegawa, T., Takizawa, M. & Tanida, S. A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol. 29, 319–322 (1983).
Tamura, T., Ishida, Y. & Suzuki, K-I. Descriptions of Actinoplanes ianthinogenes nom. rev. and Actinoplanes octamycinicus corrig. comb. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 61, 2916–2921 (2011).
Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids, Microbial ID, Inc.: Newark, Delaware, (1990).
Minnikin, D. E. et al. An integrated procedure for the extraction of bacterial isoprenoid quinines and polar lipids. J. Microbiol. Methods 2, 233–241 (1984).
Hamada, M., Iino, T., Iwami, T., Harayama, S., Tamura, T. & Suzuki, K. Mobilicoccus pelagius gen. nov., sp. nov. and Piscicoccus intestinalis gen. nov., sp. nov., two new members of the family Dermatophilaceae, and reclassification of Dermatophilus chelonae (Masters et al. 1995) as Austwickia chelonae gen. nov., comb. nov. J. Gen. Appl. Microbiol. 56, 427–436 (2010).
Tamaoka, J. & Komagata, K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128 (1984).
Saito, H. & Miura, K.-I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72, 619–629 (1963).
Hatano, K., Nishii, T. & Kasai, H. Taxonomic re-evaluation of whorl-forming Streptomyces (formerly Streptoverticillium) species by using phenotypes, DNA–DNA hybridization and sequences of gyrB, and proposal of Streptomyces luteireticuli (ex Katoh and Arai 1957) corrig., sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 53, 1519–1529 (2003).
Tamura, T. & Hatano, K. Phylogenetic analysis of the genus Actinoplanes and transfer of Actinoplanes minutisporangius Ruan et al. 1986 and ‘Actinoplanes aurantiacus’ to Cryptosporangium minutisporangium comb. nov. and Cryptosporangium aurantiacum sp. nov. Int. J. Syst. Evol. Microbiol. 51, 2119–2125 (2001).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. & Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Takahashi, K. & Nei, M. Efficiencies of fast algorithms of phylogenetic inference under the criteria of maximum parsimony, minimum evolution, and maximum likelihood when a large number of sequences are used. Mol. Biol. Evol. 17, 1251–1258 (2000).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Blin, K. et al. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41, W204–W212 (2013).
Kusunoki, S. et al. Application of colorimetric microdilution plate hybridization for rapid genetic identification of 22 Mycobacterium species. J. Clin. Microbiol. 29, 1596–1603 (1991).
Acknowledgements
This work was conducted and supported under the joint research project ‘Locally produced and Consumed’ between the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Hokuto City and the University of Yamanashi. This study was supported in part by a research grant from the Institute for Fermentation, Osaka (IFO), Japan. We are grateful to Dr Jean P Euzéby for assistance with nomenclature.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Yamamura, H., Ashizawa, H., Hamada, M. et al. Streptomyces hokutonensis sp. nov., a novel actinomycete isolated from the strawberry root rhizosphere. J Antibiot 67, 465–470 (2014). https://doi.org/10.1038/ja.2014.20
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ja.2014.20
Keywords
This article is cited by
-
Phenolic compounds as antioxidants and chemopreventive drugs from Streptomyces cellulosae strain TES17 isolated from rhizosphere of Camellia sinensis
BMC Complementary and Alternative Medicine (2018)
-
Streptomyces lutosisoli sp. nov., a novel actinomycete isolated from muddy soil
Antonie van Leeuwenhoek (2018)
-
Streptomyces flavalbus sp. nov., an actinobacterium isolated from rhizosphere of maize (Zea mays L.)
Antonie van Leeuwenhoek (2018)
-
Streptomyces euryhalinus sp. nov., a new actinomycete isolated from a mangrove forest
The Journal of Antibiotics (2017)
-
Streptomyces xiangtanensis sp. nov., isolated from a manganese-contaminated soil
Antonie van Leeuwenhoek (2017)