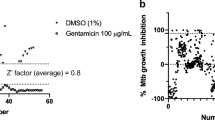
Abstract
The enzymes responsible for biotin biosynthesis in mycobacteria have been considered as potential drug targets owing to the important role in infection and cell survival that the biotin synthetic pathway plays in Mycobacterium tuberculosis. Among the enzymes that comprise mycobacterium biotin biosynthesis systems, 7,8-diaminopelargonic acid synthase (DAPAS) plays an essential role during the stationary phase in bacterial growth. In this study, compounds that inhibit mycobacterial DAPAS were screened in the virtual chemical library using an in silico structure-based drug screening (SBDS) technique, and the antimycobacterial activity of the selected compounds was validated experimentally. The DOCK–GOLD programs utilized by in silico SBDS facilitated the identification of a compound, referred to as KMD6, with potent inhibitory effects on the growth of model mycobacteria (M. smegmatis). The subsequent compound search, which was based on the structural features of KMD6, resulted in identification of three additional active compounds, designated as KMDs3, KMDs9 and KMDs10. The inhibitory effect of these compounds was comparable to that of isoniazid, which is a first-line antituberculosis drug. The high antimycobacterial activity of KMD6, KMDs9 and KMDs10 was maintained on the experiment with M. tuberculosis. Of the active compounds identified, KMDs9 would be a promising pharmacophore, owing to its long-term antimycobacterial effect and lack of cytotoxicity.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Dye, C. & Williams, B. G. The population dynamics and control of tuberculosis. Science 328, 856–861 (2010).
Lönnroth, K. et al. Tuberculosis control and elimination 2010-50: cure, care, and social development. Lancet 375, 1814–1829 (2010).
Das, P. & Horton, R. Tuberculosis–time to accelerate progress. Lancet 375, 1755–1757 (2010).
Das, P. & Horton, R. Tuberculosis–getting to zero. Lancet 386, 2231–2232 (2015).
Gandhi, N. R. et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375, 1830–1843 (2010).
Tang, J., Yam, W. C. & Chen, Z. Mycobacterium tuberculosis infection and vaccine development. Tuberculosis 98, 30–41 (2016).
Sassetti, C. M. & Rubin, E. J. Genetic requirements for mycobacterial survival during infection. Proc. Natl Acad. Sci. USA 100, 12989–12994 (2003).
Woong Park, S. et al. Evaluating the sensitivity of Mycobacterium tuberculosis to biotin deprivation using regulated gene expression. PLoS Pathog. 7, e1002264 (2011).
Dey, S., Lane, J. M., Lee, R. E., Rubin, E. J. & Sacchettini, J. C. Structural characterization of the Mycobacterium tuberculosis biotin biosynthesis enzymes 7,8-diaminopelargonic acid synthase and dethiobiotin synthetase. Biochemistry 49, 6746–6760 (2010).
Cole, S. T. et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998).
Keer, J., Smeulders, M. J., Gray, K. M. & Williams, H. D. Mutants of Mycobacterium smegmatis impaired in stationary-phase survival. Microbiology 146, 2209–2217 (2000).
Izumizono, Y., Arevalo, S., Koseki, Y., Kuroki, M. & Aoki, S. Identification of novel potential antibiotics for tuberculosis by in silico structure-based drug screening. Eur. J. Med. Chem. 46, 1849–1856 (2011).
Kinjo, T. et al. Identification of compounds with potential antibacterial activity against Mycobacterium through structure-based drug screening. J. Chem. Inf. Model. 53, 1200–1212 (2013).
Kanetaka, H. et al. Discovery of InhA inhibitors with anti-mycobacterial activity through a matched molecular pair approach. Eur. J. Med. Chem. 94, 378–385 (2015).
Koseki, Y., Kinjo, T., Kobayashi, M. & Aoki, S. Identification of novel antimycobacterial chemical agents through the in silico multi-conformational structure-based drug screening of a large-scale chemical library. Eur. J. Med. Chem. 60, 333–339 (2013).
Lang, P. T. et al. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA 15, 1219–1230 (2009).
Jones, G., Willett, P. & Glen, R. C. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 245, 43–53 (1995).
Jones, G., Willett, P., Glen, R. C., Leach, A. R. & Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 (1997).
Kobayashi, M. et al. Identification of novel potential antibiotics against Staphylococcus using structure-based drug screening targeting dihydrofolate reductase. J. Chem. Inf. Model. 54, 1242–1253 (2014).
Labute, P. LowModeMD–implicit low-mode velocity filtering applied to conformational search of macrocycles and protein loops. J. Chem. Inf. Model. 50, 792–800 (2010).
Mann, S., Marquet, A. & Ploux, O. Inhibition of 7,8-diaminopelargonic acid aminotransferase by amiclenomycin and analogues. Biochem. Soc. Trans. 33, 802–805 (2005).
Eliot, A. C., Sandmark, J., Schneider, G. & Kirsch, J. F. The dual-specific active site of 7,8-diaminopelargonic acid synthase and the effect of the R391A mutation. Biochemistry 41, 12582–12589 (2002).
Hotta, K., Kitahara, T. & Okami, Y. Studies of the mode of action of amiclenomycin. J. Antibiot. 28, 222–228 (1975).
Sandmark, J., Mann, S., Marquet, A. & Schneider, G. Structural basis for the inhibition of the biosynthesis of biotin by the antibiotic amiclenomycin. J. Biol. Chem. 277, 43352–43358 (2002).
Acknowledgements
This work was supported in part by a grant from Takeda Science Foundation to JT and a Grants-in-Aid for Young Scientists (B) (16K21226) and a Grant-in-Aid for Scientific Research (C) (26460145) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Taira, J., Morita, K., Kawashima, S. et al. Identification of a novel class of small compounds with anti-tuberculosis activity by in silico structure-based drug screening. J Antibiot 70, 1057–1064 (2017). https://doi.org/10.1038/ja.2017.106
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ja.2017.106
This article is cited by
-
Identification of novel inhibitors for mycobacterial polyketide synthase 13 via in silico drug screening assisted by the parallel compound screening with genetic algorithm-based programs
The Journal of Antibiotics (2022)
-
Improvement of the novel inhibitor for Mycobacterium enoyl-acyl carrier protein reductase (InhA): a structure–activity relationship study of KES4 assisted by in silico structure-based drug screening
The Journal of Antibiotics (2020)
-
Mimicking the human environment in mice reveals that inhibiting biotin biosynthesis is effective against antibiotic-resistant pathogens
Nature Microbiology (2019)