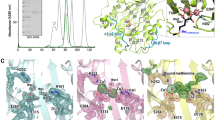
Abstract
Cytochrome c oxidase (COX) of the electron transport system is thought to be the rate-limiting step in cellular respiration and is found mutated in numerous human pathologies. Here, we employ quaternary three-dimensional (3-D) modeling to construct a model for human COX. The model was used to predict the functional consequences of amino-acid mutations based on phylogenetic conservation of amino acids together with volume and/or steric perturbations, participation in subunit–subunit interfaces and non-covalent energy loss or incompatibilities. These metrics were combined and interpreted for potential functional impact. A notable strength of the 3-D model is that it can interpret and predict the structural consequences of amino-acid variation in all 13 protein subunits. Importantly, the influence of compensatory changes can also be modeled. We examine mutations listed in the human mutation database Mitomap, and in 100 older men, and compare the results from the 3-D model against the automated MutPred web application tool. In combination, these comparisons suggest that the 3-D model predicts more functionally significant mutations than does MutPred. We conclude that the model has useful functional prediction capability but may need modification as functional data on specific mutations becomes known.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Shoubridge, E. A. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106, 46–52 (2001).
Villani, G., Greco, M., Papa, S. & Attardi, G. Low reserve of cytochrome c oxidase capacity in vivo in the respiratory chain of a variety of human cell types. J. Biol. Chem. 273, 31829–31836 (1998).
Barrientos, A., Barros, M. H., Valnot, I., Rotig, A., Rustin, P. & Tzagoloff, A. Cytochrome oxidase in health and disease. Gene 286, 53–63 (2002).
Ongwijitwat, S. & Wong-Riley, M. T. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene 360, 65–77 (2005).
Dhar, S. S., Ongwijitwat, S. & Wong-Riley, M. T. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J. Biol. Chem. 283, 3120–3129 (2008).
Osada, N. & Akashi, H. Mitochondrial–nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome C oxidase complex. Mol. Biol. Evol. 29, 337–346 (2012).
Comi, G. P., Bordoni, A., Salani, S., Franceschina, L., Sciacco, M., Prelle, A. et al. Cytochrome c oxidase subunit I microdeletion in a patient with motor neuron disease. Ann. Neurol. 43, 110–116 (1998).
Clark, K. M., Taylor, R. W., Johnson, M. A., Chinnery, P. F., Chrzanowska-Lightowlers, Z. M., Andrews, R. M. et al. An mtDNA mutation in the initiation codon of the cytochrome C oxidase subunit II gene results in lower levels of the protein and a mitochondrial encephalomyopathy. Am. J. Hum. Genet. 64, 1330–1339 (1999).
Rahman, S., Taanman, J. W., Cooper, J. M., Nelson, I., Hargreaves, I., Meunier, B. et al. A missense mutation of cytochrome oxidase subunit II causes defective assembly and myopathy. Am. J. Hum. Genet. 65, 1030–1039 (1999).
Horvath, R., Schoser, B. G., Muller-Hocker, J., Volpel, M., Jaksch, M. & Lochmuller, H. Mutations in mtDNA-encoded cytochrome c oxidase subunit genes causing isolated myopathy or severe encephalomyopathy. Neuromuscul. Disord. 15, 851–857 (2005).
Hoffbuhr, K. C., Davidson, E., Filiano, B. A., Davidson, M., Kennaway, N. G. & King, M. P. A pathogenic 15-base pair deletion in mitochondrial DNA-encoded cytochrome c oxidase subunit III results in the absence of functional cytochrome c oxidase. J. Biol. Chem. 275, 13994–14003 (2000).
Tam, E. W., Feigenbaum, A., Addis, J. B., Blaser, S., Mackay, N., Al-Dosary, M. et al. A novel mitochondrial DNA mutation in COX1 leads to strokes, seizures, and lactic acidosis. Neuropediatrics 39, 328–334 (2008).
Bratic, I. & Trifunovic, A. Mitochondrial energy metabolism and ageing. Biochim. Biophys. Acta 1797, 961–967 (2010).
Petros, J. A., Baumann, A. K., Ruiz-Pesini, E., Amin, M. B., Sun, C. Q., Hall, J. et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl Acad. Sci. USA 102, 719–724 (2005).
Diaz, F. Cytochrome c oxidase deficiency: patients and animal models. Biochim. Biophys. Acta 1802, 100–110 (2010).
Raha, S., Merante, F., Shoubridge, E., Myint, A. T., Tein, I., Benson, L. et al. Repopulation of rho0 cells with mitochondria from a patient with a mitochondrial DNA point mutation in tRNA(Gly) results in respiratory chain dysfunction. Hum. Mutat. 13, 245–254 (1999).
Baranowska, I., Jaderlund, K. H., Nennesmo, I., Holmqvist, E., Heidrich, N., Larsson, N. G. et al. Sensory ataxic neuropathy in golden retriever dogs is caused by a deletion in the mitochondrial tRNATyr gene. PLoS Genet. 5, e1000499 (2009).
Hiona, A., Sanz, A., Kujoth, G. C., Pamplona, R., Seo, A. Y., Hofer, T. et al. Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS One 5, e11468 (2010).
Dantzer, J., Moad, C., Heiland, R. & Mooney, S. MutDB services: interactive structural analysis of mutation data. Nucleic Acids Res. 33, W311–W314 (2005).
Pinto, J. R., Parvatiyar, M. S., Jones, M. A., Liang, J., Ackerman, M. J. & Potter, J. D. A functional and structural study of troponin C mutations related to hypertrophic cardiomyopathy. J. Biol. Chem. 284, 19090–19100 (2009).
Hicks, S., Wheeler, D. A., Plon, S. E. & Kimmel, M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum. Mutat. 32, 661–668 (2011).
Melvin, R. G., Katewa, S. D. & Ballard, J. W. O. A candidate complex approach to study functional mitochondrial DNA changes: sequence variation and quaternary structure modeling of Drosophila simulans cytochrome c oxidase. J. Mol. Evol. 66, 232–242 (2008).
Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K. et al. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science 269, 1069–1074 (1995).
Suga, M., Yano, N., Muramoto, K., Shinzawa-Itoh, K., Maeda, T., Yamashita, E. et al. Distinguishing between Cl− and O2(2−) as the bridging element between Fe3+ and Cu2+ in resting-oxidized cytochrome c oxidase. Acta Crystallogr. D 67, 742–744 (2011).
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Li, B., Krishnan, V. G., Mort, M. E., Xin, F., Kamati, K. K., Cooper, D. N. et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25, 2744–2750 (2009).
Ruiz-Pesini, E., Lott, M. T., Procaccio, V., Poole, J. C., Brandon, M. C., Mishmar, D. et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 35, D823–D828 (2007).
Pei, J. & Grishin, N. V. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics 23, 802–808 (2007).
Reid, B. P. O. L. M. & Sparks, R. K. Methane–methane isotropic interaction potential from total differential cross sections. J. Chem. Phys. 11, 5656 (1985).
Ratnaparkhi, G. S. & Varadarajan, R. Thermodynamic and structural studies of cavity formation in proteins suggest that loss of packing interactions rather than the hydrophobic effect dominates the observed energetics. Biochemistry 39, 12365–12374 (2000).
Holder, J. B., Bennett, A. F., Chen, J., Spencer, D. S., Byrne, M. P. & Stites, W. E. Energetics of side chain packing in staphylococcal nuclease assessed by exchange of valines, isoleucines, and leucines. Biochemistry 40, 13998–14003 (2001).
Cumming, R. G., Handelsman, D., Seibel, M. J., Creasey, H., Sambrook, P., Waite, L. et al. Cohort Profile: the Concord Health and Ageing in Men Project (CHAMP). Int. J. Epidemiol. 38, 374–378 (2009).
t Hart, L. M., Jansen, J. J., Lemkes, H. H., de Knijff, P. & Maassen, J. A. Heteroplasmy levels of a mitochondrial gene mutation associated with diabetes mellitus decrease in leucocyte DNA upon aging. Hum. Mutat. 7, 193–197 (1996).
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Colovos, C. & Yeates, T. O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519 (1993).
Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R. & Thornton, J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996).
Hooft, R. W., Vriend, G., Sander, C. & Abola, E. E. Errors in protein structures. Nature 381, 272 (1996).
Uusimaa, J., Finnila, S., Vainionpaa, L., Karppa, M., Herva, R., Rantala, H. et al. A mutation in mitochondrial DNA-encoded cytochrome c oxidase II gene in a child with Alpers–Huttenlocher-like disease. Pediatrics 111, e262–e268 (2003).
Mkaouar-Rebai, E., Ellouze, E., Chamkha, I., Kammoun, F., Triki, C. & Fakhfakh, F. Molecular–clinical correlation in a family with a novel heteroplasmic Leigh syndrome missense mutation in the mitochondrial cytochrome c oxidase III gene. J. Child Neurol. 26, 12–20 (2011).
Tiranti, V., Corona, P., Greco, M., Taanman, J. W., Carrara, F., Lamantea, E. et al. A novel frameshift mutation of the mtDNA COIII gene leads to impaired assembly of cytochrome c oxidase in a patient affected by Leigh-like syndrome. Hum. Mol. Genet. 9, 2733–2742 (2000).
Brown, M. D., Yang, C. C., Trounce, I., Torroni, A., Lott, M. T. & Wallace, D. C. A mitochondrial DNA variant, identified in Leber hereditary optic neuropathy patients, which extends the amino acid sequence of cytochrome c oxidase subunit I. Am. J. Hum. Genet. 51, 378–385 (1992).
Abu-Amero, K. K. & Bosley, T. M. Mitochondrial abnormalities in patients with LHON-like optic neuropathies. Invest. Ophthalmol. Vis. Sci. 47, 4211–4220 (2006).
Manfredi, G., Schon, E. A., Moraes, C. T., Bonilla, E., Berry, G. T., Sladky, J. T. et al. A new mutation associated with MELAS is located in a mitochondrial DNA polypeptide-coding gene. Neuromuscul. Disord. 5, 391–398 (1995).
Varlamov, D. A., Kudin, A. P., Vielhaber, S., Schroder, R., Sassen, R., Becker, A. et al. Metabolic consequences of a novel missense mutation of the mtDNA CO I gene. Hum. Mol. Genet. 11, 1797–1805 (2002).
Hamblet, N. S., Ragland, B., Ali, M., Conyers, B. & Castora, F. J. Mutations in mitochondrial-encoded cytochrome c oxidase subunits I, II, and III genes detected in Alzheimer’s disease using single-strand conformation polymorphism. Electrophoresis 27, 398–408 (2006).
Broker, S., Meunier, B., Rich, P., Gattermann, N. & Hofhaus, G. MtDNA mutations associated with sideroblastic anaemia cause a defect of mitochondrial cytochrome c oxidase. Eur. J. Biochem. 258, 132–138 (1998).
Greaves, L. C., Barron, M. J., Plusa, S., Kirkwood, T. B., Mathers, J. C., Taylor, R. W. et al. Defects in multiple complexes of the respiratory chain are present in ageing human colonic crypts. Exp. Gerontol. 45, 573–579 (2010).
Namslauer, I. & Brzezinski, P. A mitochondrial DNA mutation linked to colon cancer results in proton leaks in cytochrome c oxidase. Proc. Natl Acad. Sci. USA 106, 3402–3407 (2009).
Lucioli, S., Hoffmeier, K., Carrozzo, R., Tessa, A., Ludwig, B. & Santorelli, F. M. Introducing a novel human mtDNA mutation into the Paracoccus denitrificans COX I gene explains functional deficits in a patient. Neurogenetics 7, 51–57 (2006).
Bortot, B., Barbi, E., Biffi, S., Angelini, C., Faleschini, E., Severini, G. M. et al. Two novel cosegregating mutations in tRNAMet and COX III, in a patient with exercise intolerance and autoimmune polyendocrinopathy. Mitochondrion 9, 123–129 (2009).
Herrero-Martin, M. D., Pineda, M., Briones, P., Lopez-Gallardo, E., Carreras, M., Benac, M. et al. A new pathologic mitochondrial DNA mutation in the cytochrome oxidase subunit I (MT-CO1). Hum. Mutat. 29, E112–E122 (2008).
Wei, Y. L., Yu, C. A., Yang, P., Li, A. L., Wen, J. Y., Zhao, S. M. et al. Novel mitochondrial DNA mutations associated with Chinese familial hypertrophic cardiomyopathy. Clin. Exp. Pharmacol. Physiol. 36, 933–939 (2009).
Massa, V., Fernandez-Vizarra, E., Alshahwan, S., Bakhsh, E., Goffrini, P., Ferrero, I. et al. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am. J. Hum. Genet. 82, 1281–1289 (2008).
Wong, L. J., Dai, P., Tan, D., Lipson, M., Grix, A., Sifry-Platt, M. et al. Severe lactic acidosis caused by a novel frame-shift mutation in mitochondrial-encoded cytochrome c oxidase subunit II. Am. J. Med. Genet. 102, 95–99 (2001).
Valente, L., Piga, D., Lamantea, E., Carrara, F., Uziel, G., Cudia, P. et al. Identification of novel mutations in five patients with mitochondrial encephalomyopathy. Biochim. Biophys. Acta 1787, 491–501 (2009).
Bruno, C., Martinuzzi, A., Tang, Y., Andreu, A. L., Pallotti, F., Bonilla, E. et al. A stop-codon mutation in the human mtDNA cytochrome c oxidase I gene disrupts the functional structure of complex IV. Am. J. Hum. Genet. 65, 611–620 (1999).
Campos, Y., Garcia-Redondo, A., Fernandez-Moreno, M. A., Martinez-Pardo, M., Goda, G., Rubio, J. C. et al. Early-onset multisystem mitochondrial disorder caused by a nonsense mutation in the mitochondrial DNA cytochrome C oxidase II gene. Ann. Neurol. 50, 409–413 (2001).
Hanna, M. G., Nelson, I. P., Rahman, S., Lane, R. J., Land, J., Heales, S. et al. Cytochrome c oxidase deficiency associated with the first stop-codon point mutation in human mtDNA. Am. J. Hum. Genet. 63, 29–36 (1998).
Karadimas, C. L., Greenstein, P., Sue, C. M., Joseph, J. T., Tanji, K., Haller, R. G. et al. Recurrent myoglobinuria due to a nonsense mutation in the COX I gene of mitochondrial DNA. Neurology 55, 644–649 (2000).
Keightley, J. A., Hoffbuhr, K. C., Burton, M. D., Salas, V. M., Johnston, W. S., Penn, A. M. et al. A microdeletion in cytochrome c oxidase (COX) subunit III associated with COX deficiency and recurrent myoglobinuria. Nat. Genet. 12, 410–416 (1996).
Horvath, R., Scharfe, C., Hoeltzenbein, M., Do, B. H., Schroder, C., Warzok, R. et al. Childhood onset mitochondrial myopathy and lactic acidosis caused by a stop mutation in the mitochondrial cytochrome c oxidase III gene. J. Med. Genet. 39, 812–816 (2002).
Kollberg, G., Moslemi, A. R., Lindberg, C., Holme, E. & Oldfors, A. Mitochondrial myopathy and rhabdomyolysis associated with a novel nonsense mutation in the gene encoding cytochrome c oxidase subunit I. J. Neuropathol. Exp. Neurol. 64, 123–128 (2005).
Abu-Amero, K. K., Bosley, T. M. & Morales, J. Analysis of nuclear and mitochondrial genes in patients with pseudoexfoliation glaucoma. Mol. Vis. 14, 29–36 (2008).
Shteyer, E., Saada, A., Shaag, A., Al-Hijawi, F. A., Kidess, R., Revel-Vilk, S. et al. Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis are caused by a mutation in the COX4I2 gene. Am. J. Hum. Genet. 84, 412–417 (2009).
McFarland, R., Taylor, R. W., Chinnery, P. F., Howell, N. & Turnbull, D. M. A novel sporadic mutation in cytochrome c oxidase subunit II as a cause of rhabdomyolysis. Neuromuscul. Disord. 14, 162–166 (2004).
Marotta, R., Chin, J., Kirby, D. M., Chiotis, M., Cook, M. & Collins, S. J. Novel single base pair COX III subunit deletion of mitochondrial DNA associated with rhabdomyolysis. J. Clin. Neurosci. 18, 290–292 (2011).
Elliott, H. R., Samuels, D. C., Eden, J. A., Relton, C. L. & Chinnery, P. F. Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet. 83, 254–260 (2008).
Schaefer, A. M., McFarland, R., Blakely, E. L., He, L., Whittaker, R. G., Taylor, R. W. et al. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 63, 35–39 (2008).
Yoshikawa, S., Muramoto, K. & Shinzawa-Itoh, K. Proton-pumping mechanism of cytochrome C oxidase. Annu. Rev. Biophys. 40, 205–223 (2011).
Acknowledgements
We thank the investigators involved with the CHAMP study, including Professor Robert Cumming, Associate Professor Fiona Blyth, Associate Professor Vasikaran Naganathan, Professor David Handelsman and Melisa Litchfield for the kind provision of DNA and epidemiological data. We also thank members of the Ballard lab group for comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Horan, M., Rumbley, J., Melvin, R. et al. Quaternary protein modeling to predict the function of DNA variation found in human mitochondrial cytochrome c oxidase. J Hum Genet 58, 127–134 (2013). https://doi.org/10.1038/jhg.2012.144
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2012.144