Abstract
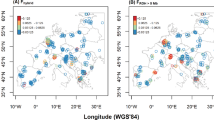
Many studies have tackled the existence of a genetic barrier in the Strait of Gibraltar between Iberian and North African populations, often with controversial conclusions. Here, we address this issue using a collection of Western Mediterranean populations and two dimensionality reduction methods: principal component analysis (PCA) and spatial PCA (sPCA). Our four different data sets consisted of (i) 16 polymorphic Alu insertions in 12 populations; (ii) 35 single-nucleotide polymorphisms in 13 populations; (iii) 13 short tandem repeats in 11 populations; and (iv) all 64 markers in 9 populations. In all PCA plots, South European and North African samples were visually distinguishable along the first PC. Several smaller clusters were also identifiable, especially on the African side of our geographical setting. sPCA indicated a single global structure for each of the marker sets and no local structures. These results are more compatible with a clinal distribution of allele frequencies rather than with abrupt changes, suggesting that isolation-by-distance, rather than a barrier to gene flow, is a more likely mechanism of genetic differentiation in the Western Mediterranean. An alternative/complementary explanation is progressive introgression from North African to Southwestern European populations.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. The history and geography of human genes, (Princeton University Press: Princeton, NJ, USA, 1994).
Botigué, L. R., Henn, B. M., Gravel, S., Maples, B. K., Gignoux, C. R., Corona, E. et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc. Natl Acad. Sci. USA 110, 11791–11796 (2013).
González-Pérez, E., Via, M., Esteban, E., López-Alomar, A., Mazieres, S., Harich, N. et al. Alu insertions in the Iberian Peninsula and North West Africa - genetic boundaries or melting pot? Coll. Antropol. 27, 491–500 (2003).
Barbujani, G., Pilastro, A., De Domenico, S. & Renfrew, C. Genetic variation in North Africa and Eurasia: Neolithic demic difusion vs. Paleolithic colonisation. Am. J. Phys. Anthropol. 95, 137–154 (1994).
Kandil, M., Moral, P., Esteban, E., Autori, L., Mameli, G. E., Zaoui, D. et al. Red cell enzyme polymorphisms in Moroccans and southern Spaniards: new data for the genetic history of the western Mediterranean. Hum. Biol. 71, 791–802 (1999).
Plaza, S., Calafell, F., Helal, A., Bouzerna, N., Lefranc, G., Bertranpetit, J. et al. Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean. Ann. Hum. Genet. 67, 312–328 (2003).
Bahri, R., Esteban, E., Moral, P. & Chaabani, H. New insights into the genetic history of Tunisians: data from Alu insertion and apolipoprotein E gene polymorphisms. Ann. Hum. Biol. 35, 22–33 (2008).
González-Pérez, E., Esteban, E., Via, M., Gayà-Vidal, M., Athanasiadis, G., Dugoujon, J. M. et al. Population relationships in the Mediterranean revealed by autosomal genetic data (Alu and Alu/STR compound systems). Am. J. Phys. Anthropol. 141, 430–439 (2010).
Bosch, E., Calafell, F., Pérez-Lezaun, A., Comas, D., Mateu, E. & Bertranpetit, J. Population history of North Africa: Evidence from classical genetic markers. Hum. Biol. 69, 295–311 (1997).
Comas, D., Calafell, F., Benchemsi, N., Helal, A., Lefranc, G., Stoneking, M. et al. Alu insertion polymorphisms in NW Africa and the Iberian Peninsula: evidence for a strong genetic boundary through the Gibraltar Straits. Hum. Genet. 107, 312–319 (2000).
Harich, N., Esteban, E., Chafik, A., López-Alomar, A., Vona, G. & Moral, P. Classical polymorphisms in Berbers from Moyen Atlas (Morocco): genetics, geography, and historical evidence in the Mediterranean peoples. Ann. Hum. Biol. 29, 473–487 (2002).
Athanasiadis, G., González-Pérez, E., Esteban, E., Dugoujon, J. M., Stoneking, M. & Moral, P. The Mediterranean Sea as a barrier to gene flow: evidence from variation in and around the F7 and F12 genomic regions. BMC Evol. Biol. 10, 84 (2010).
Jombart, T., Devillard, S., Dufour, A. B. & Pontier, D. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101, 92–103 (2008).
Moran, P. The interpretation of statistical maps. J. R. Stat. Soc. Ser. B 10, 243–251 (1948).
Athanasiadis, G., Esteban, E., Gayà-Vidal, M., Dugoujon, J. M., Moschonas, N., Chaabani, H. et al. Different evolutionary histories of the Coagulation Factor VII gene in human populations? Ann. Hum. Genet. 74, 34–45 (2010).
Mantel, N. Detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220 (1967).
Chessel, D., Dufour, A. B. & Thioulouse, J. The ade4 package- I: one-table methods. R News 4, 5–10 (2004).
Jombart, T. & Ahmed, I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011).
Bar-Yosef, O. The upper paleolithic revolution. Annu. Rev. Antropol. 31, 363–393 (2002).
Xing, J., Watkins, W.S., Witherspoon, D.J., Zhang, Y., Guthery, S.L., Thara, R. et al. Fine-scaled human genetic structure revealed by SNP microarrays. Genome Res. 19, 815–825 (2009).
Acknowledgements
The authors would like to thank two anonymous reviewers for their useful and constructive comments on the manuscript. This study has been financially supported by the CGL2008-03955 and CGL2011-27866 projects of the Spanish Ministerio de Educación y Ciencia, as well as the 2005SGR00252 project of the Generalitat de Catalunya. The work of GA has been financed by an FPU grant from the Ministerio de Educación y Ciencia (reference: AP2005-4425). The authors would also like to thank all the donors for providing blood samples and the collaborators for collecting them: F Luna, MS Mesa and C Rodríguez (Spanish samples); N Harich and M Kandil (Moroccan samples); H Chaabani (Tunisian sample), JM Dugoujon (North African and French samples); and, two colleagues: M Gayà-Vidal for helping in the SNP genotyping and E González-Pérez for providing the PAI dataset. Also thanks to Professor M Stoneking for his advice, technical and material support during GA’s visit at the Max Planck Institute for Evolutionary Anthropology in Leipzig (Germany), where most of the STR genotyping was carried out. Finally, the authors would like to thank Professor WH Stone and ME Weale for reviewing and discussing the material presented here.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Athanasiadis, G., Moral, P. Spatial principal component analysis points at global genetic structure in the Western Mediterranean. J Hum Genet 58, 762–765 (2013). https://doi.org/10.1038/jhg.2013.94
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2013.94
Keywords
This article is cited by
-
A survey of sub-Saharan gene flow into the Mediterranean at risk loci for coronary artery disease
European Journal of Human Genetics (2017)