Abstract
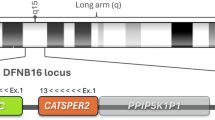
With homozygosity mapping we have identified two large homozygous regions on chromosome 3q13.11–q13.31 and chromosome 19p13.3–q31.32 in a large Pakistani family suffering from autosomal recessive nonsyndromic hearing impairment (arNSHI). The region on chromosome 19 overlaps with the previously described deafness loci DFNB15, DFNB72 and DFNB95. Mutations in GIPC3 have been shown to underlie the nonsyndromic hearing impairment linked to these loci. Sequence analysis of all exons and exon–intron boundaries of GIPC3 revealed a homozygous canonical splice site mutation, c.226-1G>T, in GIPC3. This is the first mutation described in GIPC3 that affects splicing. The c.226-1G>T mutation is located in the acceptor splice site of intron 1 and is predicted to affect the normal splicing of exon 2. With a minigene assay it was shown to result in the use of an alternative acceptor site in exon 2, resulting in a frameshift and a premature stop codon. This study expands the mutational spectrum of GIPC3 in arNSHI.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Shafique S, Siddiqi S, Schraders M, Oostrik J, Ayub H, Bilal A et al. Genetic spectrum of autosomal recessive non-syndromic hearing loss in pakistani families. PLoS ONE 2014; 9: e100146.
Sambrook J, Fritsch EF & Maniatis T Analysis and Cloning of Eukaryotic Genomic DNA. Molecular Cloning: A Laboratory Manual. (Cold Spring Harbour Laboratory Press, New York, 1989).
Seelow D, Schuelke M, Hildebrandt F & Nurnberg P HomozygosityMapper—an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009; 37: W593–W599.
Ain Q, Nazli S, Riazuddin S, Jaleel AU, Riazuddin SA, Zafar AU et al. The autosomal recessive non-syndromic deafness locus DFNB72 is located on chromosome 19p13.3. Hum. Genet. 2007; 122: 445–450.
Charizopoulou N, Lelli A, Schraders M, Ray K, Hildebrand MS, Ramesh A et al. Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat. Commun. 2011; 2: 201.
Chen A, Wayne S, Bell A, Ramesh A, Srisailapathy CR, Scott DA et al. New gene for autosomal recessive non-syndromic hearing loss maps to either chromosome 3q or 19p. Am. J. Med. Genet. 1997; 71: 467–471.
Rehman AU, Gul K, Morell RJ, Lee K, Ahmed ZM, Riazuddin S et al. Mutations of GIPC3 cause nonsyndromic hearing loss DFNB72 but not DFNB81 that also maps to chromosome 19p. Hum. Genet. 2011; 130: 759–765.
Santos RL, Hassan MJ, Sikandar S, Lee K, Ali G, Martin PE Jr. et al. DFNB68, a novel autosomal recessive non-syndromic hearing impairment locus at chromosomal region 19p13.2. Hum. Genet. 2006; 120: 85–92.
Diaz-Horta O, Duman D, Foster J 2nd, Sirmaci A, Gonzalez M, Mahdieh N et al. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS ONE 2012; 7: e50628.
Ramzan K, Al-Owain M, Allam R, Berhan A, Abuharb G, Taibah K et al. Homozygosity mapping identifies a novel GIPC3 mutation causing congenital nonsyndromic hearing loss in a Saudi family. Gene 2013; 521: 195–199.
Sirmaci A, Edwards YJ, Akay H & Tekin M Challenges in whole exome sequencing: an example from hereditary deafness. PLoS ONE 2012; 7: e32000.
Acknowledgements
We are thankful to the Higher Education Commission, Pakistan for providing Post Doc scholarship to SS to conduct this study in the group of Prof. H. Kremer in the Netherlands. This work was financially supported by a grant from ZorgOnderzoek Nederland / Medische Wetenschappen (40-00812-98-09047, to H.K.).
Web resources. The URLs for the data presented herein are as follows: The Hereditary Hearing loss Homepage, http://hereditaryhearingloss.org/. HomozygosityMapper, http://www.homozygositymapper.org/. Polymorphism Phenotyping (Polyphen-2), http://genetics.bwh.harvard.edu/pph2/. Sorting Intolerant from Tolerant (SIFT), http://sift.jcvi.org/. UCSC Human Genome Database, Build hg19, http://www.genome.ucsc.edu/. Exome variant Server, http://evs.gs.washington.edu/EVS/. 1000 Genome projects, http://browser.1000genomes.org/. National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
Author contributions: SS and JO performed the experiments, SM and MI provided the family for the study. SS did the data analysis. HK provided all reagents/software. SS wrote the paper. RQ, AM, HK and MS supervised the work and critically read the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Siddiqi, S., Ismail, M., Oostrik, J. et al. A canonical splice site mutation in GIPC3 causes sensorineural hearing loss in a large Pakistani family. J Hum Genet 59, 683–686 (2014). https://doi.org/10.1038/jhg.2014.86
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2014.86
This article is cited by
-
Low incidence of GIPC3 variants among the prelingual hearing impaired from southern India
Journal of Genetics (2020)