Abstract
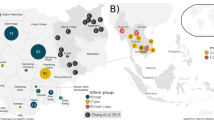
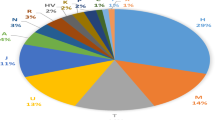
The complete mitochondrial genome of one 700-year-old individual found in Tashkurgan, Xinjiang was target enriched and sequenced in order to shed light on the population history of Tashkurgan and determine the phylogenetic relationship of haplogroup U5a. The ancient sample was assigned to a subclade of haplogroup U5a2a1, which is defined by two rare and stable transversions at 16114A and 13928C. Phylogenetic analysis shows a distribution pattern for U5a2a that is indicative of an origin in the Volga–Ural region and exhibits a clear eastward geographical expansion that correlates with the pastoral culture also entering the Eurasian steppe. The haplogroup U5a2a present in the ancient Tashkurgan individual reveals prehistoric migration in the East Pamir by pastoralists. This study shows that studying an ancient mitochondrial genome is a useful approach for studying the evolutionary process and population history of Eastern Pamir.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Museum, X. U. A. R. The investigation of Paleolithic Age ruins in Jirzanka of Tashkurgan Tajik autonomous county. Xinjiang Relics 1, 6 (1985).
Malyarchuk, B., Derenko, M., Grzybowski, T., Perkova, M., Rogalla, U., Vanecek, T. et al. The peopling of Europe from the mitochondrial haplogroup U5 perspective. PloS ONE 5, e10285 (2010).
Cui, Y., Li, C., Gao, S., Xie, C. & Zhou, H. Early Eurasian migration traces in the Tarim Basin revealed by mtDNA polymorphisms. Am. J. Phys. Anthropol. 142, 558–564 (2010).
Wu, Y. On the Bronze Culture of Xiabandi Cemetery in Kashi, Xinjiang. Western Reg. Stud. 04, 36–44 (2012).
Li, H., Zhao, X., Zhao, Y., Li, C., Si, D., Zhou, H. et al. Genetic characteristics and migration history of a bronze culture population in the West Liao-River valley revealed by ancient DNA. J. Hum. Genet. 56, 815–822 (2011).
Cui, Y., Lindo, J., Hughes, C. E., Johnson, J. W., Hernandez, A. G., Kemp, B. M. et al. Ancient DNA analysis of mid-holocene individuals from the Northwest Coast of North America reveals different evolutionary paths for mitogenomes. PloS ONE 8, e66948 (2013).
Lindgreen, S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes 5, 337 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Wei, Z., Wang, W., Hu, P., Lyon, G. J. & Hakonarson, H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 39, e132 (2011).
Jonsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. & Orlando, L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013).
Bandelt, H. J., Forster, P. & Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Fan, L & Yao, Y. G. MitoTool: a web server for the analysis and retrieval of human mitochondrial DNA sequence variations. Mitochondrion 11, 351–356 (2011).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 9, 772 (2012).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003).
Atkinson, Q. D., Gray, R. D. & Drummond, A. J. mtDNA variation predicts population size in humans and reveals a major southern Asian chapter in human prehistory. Mol. Biol. Evol. 25, 468–474 (2008).
Atkinson, Q. D., Gray, R. D. & Drummond, A. J. Bayesian coalescent inference of major human mitochondrial DNA haplogroup expansions in Africa. Proc. Biol. Sci. 276, 367–373 (2009).
Peng, M.-S. & Zhang, Y.-P. Inferring the population expansions in peopling of Japan. PloS ONE 6, e21509 (2011).
Excoffier, L., Estoup, A. & Cornuet, J. M. Bayesian analysis of an admixture model with mutations and arbitrarily linked markers. Genetics 169, 1727–1738 (2005).
Fan, L. & Yao, Y. G. An update to MitoTool: using a new scoring system for faster mtDNA haplogroup determination. Mitochondrion 13, 360–363 (2013).
Saillard, J., Forster, P., Lynnerup, N., Bandelt, H. J. & Norby, S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am. J. Hum. Genet. 67, 718–726 (2000).
Soares, P., Ermini, L., Thomson, N., Mormina, M., Rito, T., Rohl, A. et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 84, 740–759 (2009).
Cooper, A. & Poinar, H. N. Ancient DNA: do it right or not at all. Science 289, 1139 (2000).
Sawyer, S., Krause, J., Guschanski, K., Savolainen, V. & Paabo, S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PloS ONE 7, e34131 (2012).
Behar, D. M., van Oven, M., Rosset, S., Metspalu, M., Loogvali, E. L., Silva, N. M. et al. A "Copernican" reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90, 675–684 (2012).
Quintana-Murci, L., Chaix, R., Wells, R. S., Behar, D. M., Sayar, H., Scozzari, R. et al. Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am. J. Hum. Genet. 74, 827–845 (2004).
Comas, D., Calafell, F., Mateu, E., Perez-Lezaun, A., Bosch, E., Martinez-Arias, R. et al. Trading genes along the silk road: mtDNA sequences and the origin of central Asian populations. Am. J. Hum. Genet. 63, 1824–1838 (1998).
Ingman, M. & Gyllensten, U. Rate variation between mitochondrial domains and adaptive evolution in humans. Hum. Mol. Genet. 16, 2281–2287 (2007).
Irwin, J. A., Ikramov, A., Saunier, J., Bodner, M., Amory, S., Rock, A. et al. The mtDNA composition of Uzbekistan: a microcosm of Central Asian patterns. Int. J. Legal Med. 124, 195–204 (2010).
Malyarchuk, B. A. & Derenko, M. V. Mitochondrial DNA variability in Russians and Ukrainians: implication to the origin of the Eastern Slavs. Ann. Hum. Genet. 65, 63–78 (2001).
Malyarchuk, B. A., Grzybowski, T., Derenko, M. V., Czarny, J., Wozniak, M. & Miscicka-Sliwka, D. Mitochondrial DNA variability in Poles and Russians. Ann. Hum. Genet. 66, 261–283 (2002).
Pakendorf, B., Wiebe, V., Tarskaia, L. A., Spitsyn, V. A., Soodyall, H., Rodewald, A. et al. Mitochondrial DNA evidence for admixed origins of central Siberian populations. Am. J. Phys. Anthropol. 120, 211–224 (2003).
Melchior, L., Lynnerup, N., Siegismund, H. R., Kivisild, T. & Dissing, J. Genetic diversity among ancient Nordic populations. PloS ONE 5, e11898 (2010).
Bramanti, B., Thomas, M. G., Haak, W., Unterlaender, M., Jores, P., Tambets, K. et al. Genetic discontinuity between local hunter-gatherers and central Europe's first farmers. Science 326, 137–140 (2009).
Bermisheva, M., Tambets, K., Villems, R. & Khusnutdinova, E. [Diversity of mitochondrial DNA haplotypes in ethnic populations of the Volga-Ural region of Russia]. Mol. Biol. (Mosk.) 36, 990–1001 (2002).
Malyarchuk, B., Derenko, M., Grzybowski, T., Lunkina, A., Czarny, J., Rychkov, S. et al. Differentiation of mitochondrial DNA and Y chromosomes in Russian populations. Hum. Biol. 76, 877–900 (2004).
Grzybowski, T., Malyarchuk, B. A., Derenko, M. V., Perkova, M. A., Bednarek, J. & Wozniak, M. Complex interactions of the Eastern and Western Slavic populations with other European groups as revealed by mitochondrial DNA analysis. Forensic Sci. Int. Genet. 1, 141–147 (2007).
Keyser, C., Bouakaze, C., Crubezy, E., Nikolaev, V. G., Montagnon, D., Reis, T. et al. Ancient DNA provides new insights into the history of south Siberian Kurgan people. Hum. Genet. 126, 395–410 (2009).
Chernykh, E. N. Formation of the Eurasian "Steppe Belt" of stockbreeding cultures: viewed through the prism of archaeometallurgy and radiocarbon dating. Archaeol Ethnol Anthropol Eurasia 35, 36–53 (2008).
Mei, J. & Colin, S. The existence of Andronovo cultural influence in Xinjiang during the 2nd millennium BC. Antiquity 73, 570–578 (1999).
Guo, W. Archaeological Research on the Societies of the Late Prehistoric Xinjiang, (Shanghai Shiji Press, Shanghai, China, 2012).
Johnson, J. W. The Scythian: his rise and fall. J. Hist. Ideas 20, 250–257 (1959).
Ricaut, F. X., Keyser-Tracqui, C., Cammaert, L., Crubezy, E. & Ludes, B. Genetic analysis and ethnic affinities from two Scytho-Siberian skeletons. Am. J. Phys. Anthropol. 123, 351–360 (2004).
Han, K. Anceint Race Study on the Silk Road, (Xijiang People's Publishing House, Xinjiang, China, 1993).
Hey, J. On the number of New World founders: a population genetic portrait of the peopling of the Americas. PLoS Biol. 3, e193 (2005).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 41472024 and 31301025).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Ning, C., Gao, S., Deng, B. et al. Ancient mitochondrial genome reveals trace of prehistoric migration in the east Pamir by pastoralists. J Hum Genet 61, 103–108 (2016). https://doi.org/10.1038/jhg.2015.128
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2015.128
This article is cited by
-
Genetic kinship and admixture in Iron Age Scytho-Siberians
Human Genetics (2019)
-
Mitochondrial genomes uncover the maternal history of the Pamir populations
European Journal of Human Genetics (2018)
-
Cannabis in Eurasia: origin of human use and Bronze Age trans-continental connections
Vegetation History and Archaeobotany (2017)