Abstract
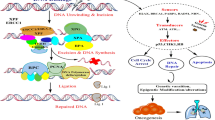
Genetic factors are important in lung cancer, but as most lung cancers are sporadic, little is known about inherited genetic factors. We identified a three-generation family with suspected autosomal dominant inherited lung cancer susceptibility. Sixteen individuals in the family had lung cancer. To identify the gene(s) that cause lung cancer in this pedigree, we extracted DNA from the peripheral blood of three individuals and from the blood of one cancer-free control family member and performed whole-exome sequencing. We identified 41 alterations in 40 genes in all affected family members but not in the unaffected member. These were considered candidate mutations for familial lung cancer. Next, to identify somatic mutations and/or inherited alterations in these 40 genes among sporadic lung cancers, we performed exon target enrichment sequencing using 192 samples from sporadic lung cancer patients. We detected somatic ‘candidate’ mutations in multiple sporadic lung cancer samples; MAST1, CENPE, CACNB2 and LCT were the most promising candidate genes. In addition, the MAST1 gene was located in a putative cancer-linked locus in the pedigree. Our data suggest that several genes act as oncogenic drivers in this family, and that MAST1 is most likely to cause lung cancer.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Nagy, R., Sweet, K. & Eng, C. Highly penetrant hereditary cancer syndromes. Oncogene 23, 6445–6470 (2004).
Amos, C. I., Pinney, S. M., Li, Y., Kupert, E., Lee, J., de Andrade, M. A. et al. A susceptibility locus on chromosome 6q greatly increases lung cancer risk among light and never smokers. Cancer Res. 70, 2359–2367 (2010).
Hung, R. J., McKay, J. D., Gaborieau, V., Boffetta, P., Hashibe, M., Zaridze, D. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 452, 633–637 (2008).
Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004).
Riely, G. J., Marks, J. & Pao, W. KRAS mutations in non-small cell lung cancer. Proc. Am. Thorac. Soc. 6, 201–205 (2009).
Yamamoto, H., Higasa, K., Sakaguchi, M., Shien, K., Soh, J., Ichimura, K. et al. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J. Natl Cancer Inst. 106, djt338 (2014).
Soda, M., Choi, Y. L., Enomoto, M., Takada, S., Yamashita, Y., Ishikawa, S. et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007).
Liu, Y., Liu, P., Wen, W., James, M. A., Wang, Y., Bailey-Wilson, J. E. et al. Haplotype and cell proliferation analyses of candidate lung cancer susceptibility genes on chromosome 15q24-25.1. Cancer Res. 69, 7844–7850 (2009).
Liu, P., Vikis, H. G., Wang, D., Lu, Y., Wang, Y., Schwartz, A. G. et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J. Natl Cancer Inst. 100, 1326–1330 (2008).
Fang, S., Pinney, S. M., Bailey-Wilson, J. E., de Andrade, M. A., Li, Y., Kupert, E. et al. Ordered subset analysis identifies loci influencing lung cancer risk on chromosomes 6q and 12q. Cancer Epidemiol. Biomarkers Prev. 19, 3157–3166 (2010).
Bailey-Wilson, J. E., Amos, C. I., Pinney, S. M., Petersen, G. M., de Andrade, M., Wiest, J. S. et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am. J. Hum. Genet. 75, 460–474 (2004).
You, M., Wang, D., Liu, P., Vikis, H., James, M., Lu, Y. et al. Fine mapping of chromosome 6q23-25 region in familial lung cancer families reveals RGS17 as a likely candidate gene. Clin. Cancer Res. 15, 2666–2674 (2009).
Liu, P., Vikis, H. G., Lu, Y., Wang, Y., Schwartz, A. G., Pinney, S. M. et al. Cumulative effect of multiple loci on genetic susceptibility to familial lung cancer. Cancer Epidemiol. Biomarkers Prev. 19, 517–524 (2010).
Wang, Y., Kuan, P. J., Xing, C., Cronkhite, J. T., Torres, F., Rosenblatt, R. L. et al. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am. J. Hum. Genet. 84, 52–59 (2009).
McKenna, A., Hanna, M. & Banks, E. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
DePristo, M. A., Banks, E., Poplin, R., Garimella, K. V., Maguire, J. R., Hartl, C. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., del Angel, G., Levy-Moonshine, A. et al. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 11, 1011.10.1–11.10.33 (2013).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Tsai, M.-F., Lin, Y.-J., Cheng, Y.-C., Lee, K.-H., Huang, C.-C., Chen, Y.-T. et al. PrimerZ: streamlined primer design for promoters, exons and human SNPs. Nucleic Acids Res. 35, W63–W65 (2007).
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R. & Leunissen, J. A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74 (2007).
Christensen, J. G., Burrows, J. & Salgia, R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 225, 1–26 (2005).
Davis, I. J., McFadden, A. W., Zhang, Y., Coxon, A., Burgess, T. L., Wagner, A. J. et al. Identification of the receptor tyrosine kinase c-Met and its ligand, hepatocyte growth factor, as therapeutic targets in clear cell sarcoma. Cancer Res. 70, 639–645 (2010).
Pao, W. & Girard, N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 12, 175–180 (2011).
Amos, C. I., Wu, X., Broderick, P., Gorlov, I. P., Gu, J., Eisen, T. et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 40, 616–622 (2008).
McKay, J. D., Hung, R. J., Gaborieau, V., Boffetta, P., Chabrier, A., Byrnes, G. et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 40, 1404–1406 (2008).
Wang, Y., Broderick, P., Webb, E., Wu, X., Vijayakrishnan, J., Matakidou, A. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 40, 1407–1409 (2008).
Valiente, M., Andrés-Pons, A., Gomar, B., Torres, J., Gil, A., Tapparel, C. et al. Binding of PTEN to specific PDZ domains contributes to PTEN protein stability and phosphorylation by microtubule-associated serine/threonine kinases. J. Biol. Chem. 280, 28936–28943 (2005).
Robinson, D. R., Kalyana-Sundaram, S., Wu, Y.-M., Shankar, S., Cao, X., Ateeq, B. et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 17, 1646–1651 (2011).
Wood, K. W., Lad, L., Luo, L., Qian, X., Knight, S. D., Nevins, N. et al. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl Acad. Sci. USA 107, 5839–5844 (2010).
Mirzaa, G. M., Vitre, B., Carpenter, G., Abramowicz, I., Gleeson, J. G., Paciorkowski, A. R. et al. Mutations in CENPE define a novel kinetochore-centromeric mechanism for microcephalic primordial dwarfism. Hum. Genet. 133, 1023–1039 (2014).
Bácsi, K., Hitre, E., Kósa, J. P., Horváth, H., Lazáry, A., Lakatos, P. L. et al. Effects of the lactase 13910 C/T and calcium-sensor receptor A986S G/T gene polymorphisms on the incidence and recurrence of colorectal cancer in Hungarian population. BMC Cancer 8, 317 (2008).
Acknowledgements
This work was supported by the Ministry of Health, Labor and Welfare of Japan (KY); the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (B) grant number 25293084 (KY), Challenging Exploratory Research grant number 25550033 (KY), Grant-in-Aid for Scientific Research (C) grant number 25462180 (TM) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website .
Supplementary information
Rights and permissions
About this article
Cite this article
Tomoshige, K., Matsumoto, K., Tsuchiya, T. et al. Germline mutations causing familial lung cancer. J Hum Genet 60, 597–603 (2015). https://doi.org/10.1038/jhg.2015.75
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2015.75
This article is cited by
-
Different driver gene mutations in patients with synchronous multiple primary lung cancers: a case report
Journal of Cardiothoracic Surgery (2020)
-
Unusual synchronous double primary treatment-naïve lung adenocarcinoma harboring T790M and L858R mutations in early-stage lung cancer
World Journal of Surgical Oncology (2019)
-
Identification of susceptibility pathways for the role of chromosome 15q25.1 in modifying lung cancer risk
Nature Communications (2018)
-
QuaDMutEx: quadratic driver mutation explorer
BMC Bioinformatics (2017)
-
Genetic susceptibility variants for lung cancer: replication study and assessment as expression quantitative trait loci
Scientific Reports (2017)