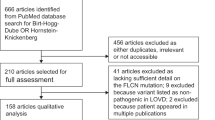
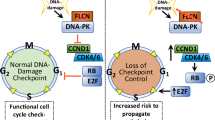
Abstract
Pathogenic germline mutations in the folliculin (FLCN) tumor suppressor gene predispose to Birt–Hogg–Dubé (BHD) syndrome, a rare disease characterized by the development of cutaneous hamartomas (fibrofolliculomas), multiple lung cysts, spontaneous pneumothoraces and renal cell cancer. In this study, we report the identification of 13 variants and three polymorphisms in the FLCN gene in 143 Danish patients or families with suspected BHD syndrome. Functional mini-gene splicing analysis revealed that two intronic variants (c.1062+2T>G and c.1177-5_1177-3del) introduced splicing aberrations. Eleven families exhibited the c.1062+2T>G mutation. Combined single nucleotide polymorphism array-haplotype analysis showed that these families share a 3-Mb genomic fragment containing the FLCN gene, revealing that the c.1062+2T>G mutation is a Danish founder mutation. On the basis of in silico prediction and functional splicing assays, we classify the 16 identified variants in the FLCN gene as follows: nine as pathogenic, one as likely pathogenic, three as likely benign and three as polymorphisms. In conclusion, the study describes the FLCN mutation spectrum in Danish BHD patients, and contributes to a better understanding of BHD syndrome and management of BHD patients.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Birt, A. R., Hogg, G. R. & Dube, W. J. Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch. Dermatol. 113, 1674–1677 (1977).
Khoo, S. K., Bradley, M., Wong, F. K., Hedblad, M. A., Nordenskjold, M. & Teh, B. T. Birt–Hogg–Dube syndrome: mapping of a novel hereditary neoplasia gene to chromosome 17p12-q11.2. Oncogene 20, 5239–5242 (2001).
Nickerson, M. L., Warren, M. B., Toro, J. R., Matrosova, V., Glenn, G., Turner, M. L. et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt–Hogg–Dube syndrome. Cancer Cell. 2, 157–164 (2002).
Schmidt, L. S. & Linehan, W. M. Molecular genetics and clinical features of Birt–Hogg–Dube syndrome. Nat. Rev. Urol. 12, 558–569 (2015).
Pavlovich, C. P., Walther, M. M., Eyler, R. A., Hewitt, S. M., Zbar, B., Linehan, W. M. et al. Renal tumors in the Birt–Hogg–Dube syndrome. Am. J. Surg. Pathol. 26, 1542–1552 (2002).
Houweling, A. C., Gijezen, L. M., Jonker, M. A., van Doorn, M. B., Oldenburg, R. A., van Spaendonck-Zwarts, K. Y. et al. Renal cancer and pneumothorax risk in Birt–Hogg–Dube syndrome; an analysis of 115 FLCN mutation carriers from 35 BHD families. Br. J. Cancer 105, 1912–1919 (2011).
Schmidt, L. S., Nickerson, M. L., Warren, M. B., Glenn, G. M., Toro, J. R., Merino, M. J. et al. Germline BHD-mutation spectrum and phenotype analysis of a large cohort of families with Birt–Hogg–Dube syndrome. Am. J. Hum. Genet. 76, 1023–1033 (2005).
Toro, J. R., Wei, M. H., Glenn, G. M., Weinreich, M., Toure, O., Vocke, C. et al. BHD mutations, clinical and molecular genetic investigations of Birt–Hogg–Dube syndrome: a new series of 50 families and a review of published reports. J. Med. Genet. 45, 321–331 (2008).
Menko, F. H., van Steensel, M. A., Giraud, S., Friis-Hansen, L., Richard, S., Ungari, S. et al. Birt–Hogg–Dube syndrome: diagnosis and management. Lancet Oncol. 10, 1199–1206 (2009).
Schmidt, L. S. & Linehan, W. M. Clinical features, genetics and potential therapeutic approaches for Birt–Hogg–Dube syndrome. Expert Opin. Orphan. Drugs 3, 15–29 (2015).
Leter, E. M., Koopmans, A. K., Gille, J. J., van Os, T. A., Vittoz, G. G., David, E. F. et al. Birt–Hogg–Dube syndrome: clinical and genetic studies of 20 families. J. Invest. Dermatol. 128, 45–49 (2008).
Woodward, E. R., Ricketts, C., Killick, P., Gad, S., Morris, M. R., Kavalier, F. et al. Familial non-VHL clear cell (conventional) renal cell carcinoma: clinical features, segregation analysis, and mutation analysis of FLCN. Clin. Cancer Res. 14, 5925–5930 (2008).
Dandanell, M., Friis-Hansen, L., Sunde, L., Nielsen, F. C. & Hansen, T. V. Identification of 3 novel VHL germ-line mutations in Danish VHL patients. BMC Med. Genet. 13, 54 (2012).
Jonson, L., Ahlborn, L. B., Steffensen, A. Y., Djursby, M., Ejlertsen, B., Timshel, S. et al. Identification of six pathogenic RAD51C mutations via mutational screening of 1228 Danish individuals with increased risk of hereditary breast and/or ovarian cancer. Breast Cancer Res. Treat. 155, 215–222 (2016).
Tavtigian, S. V., Deffenbaugh, A. M., Yin, L., Judkins, T., Scholl, T., Samollow, P. B. et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 43, 295–305 (2006).
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Ng, P. C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Shapiro, M. B. & Senapathy, P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 15, 7155–7174 (1987).
Pertea, M., Lin, X. & Salzberg, S. L. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 29, 1185–1190 (2001).
Reese, M. G., Eeckman, F. H., Kulp, D. & Haussler, D. Improved splice site detection in genie. J. Comput. Biol. 4, 311–323 (1997).
Yeo, G. & Burge, C. B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11, 377–394 (2004).
Desmet, F. O., Hamroun, D., Lalande, M., Collod-Beroud, G., Claustres, M. & Beroud, C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 (2009).
Thery, J. C., Krieger, S., Gaildrat, P., Revillion, F., Buisine, M. P., Killian, A. et al. Contribution of bioinformatics predictions and functional splicing assays to the interpretation of unclassified variants of the BRCA genes. Eur. J. Hum. Genet. 19, 1052–1058 (2011).
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Petersen, S. M., Dandanell, M., Rasmussen, L. J., Gerdes, A. M., Krogh, L. N., Bernstein, I. et al. Functional examination of MLH1, MSH2, and MSH6 intronic mutations identified in Danish colorectal cancer patients. BMC Med. Genet. 14, 103 (2013).
Steffensen, A. Y., Dandanell, M., Jonson, L., Ejlertsen, B., Gerdes, A. M., Nielsen, F. C. et al. Functional characterization of BRCA1 gene variants by mini-gene splicing assay. Eur. J. Hum. Genet. 22, 1362–1368 (2014).
Ahlborn, L. B., Dandanell, M., Steffensen, A. Y., Jonson, L., Nielsen, F. C. & Hansen, T. V. Splicing analysis of 14 BRCA1 missense variants classifies nine variants as pathogenic. Breast Cancer Res. Treat. 150, 289–298 (2015).
Hansen, T. V., Vikesaa, J., Buhl, S. S., Rossing, H. H., Timmermans-Wielenga, V. & Nielsen, F. C. High-density SNP arrays improve detection of HER2 amplification and polyploidy in breast tumors. BMC Cancer 15, 35 (2015).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Albrechtsen, A., Sand, K. T., Moltke, I., van Overseem, H. T., Nielsen, F. C. & Nielsen, R. Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium. Genet. Epidemiol. 33, 266–274 (2009).
Benusiglio, P. R., Giraud, S., Deveaux, S., Mejean, A., Correas, J. M., Joly, D. et al. Renal cell tumour characteristics in patients with the Birt–Hogg–Dube cancer susceptibility syndrome: a retrospective, multicentre study. Orphanet. J. Rare Dis. 9, 163 (2014).
Furuya, M., Yao, M., Tanaka, R., Nagashima, Y., Kuroda, N., Hasumi, H. et al. Genetic, epidemiologic and clinicopathologic studies of Japanese Asian patients with Birt–Hogg–Dube syndrome. Clin Genet. (e-pub ahead of print 25 May 2016. doi:10.1111/cge.12807).
Khoo, S. K., Giraud, S., Kahnoski, K., Chen, J., Motorna, O., Nickolov, R. et al. Clinical and genetic studies of Birt–Hogg–Dube syndrome. J. Med. Genet. 39, 906–912 (2002).
Benhammou, J. N., Vocke, C. D., Santani, A., Schmidt, L. S., Baba, M., Seyama, K. et al. Identification of intragenic deletions and duplication in the FLCN gene in Birt–Hogg–Dube syndrome. Genes Chromosomes Cancer 50, 466–477 (2011).
Ding, Y., Zhu, C., Zou, W., Ma, D., Min, H., Chen, B. et al. FLCN intragenic deletions in Chinese familial primary spontaneous pneumothorax. Am. J. Med. Genet. A 167A, 1125–1133 (2015).
Kunogi, M., Kurihara, M., Ikegami, T. S., Kobayashi, T., Shindo, N., Kumasaka, T. et al. Clinical and genetic spectrum of Birt–Hogg–Dube syndrome patients in whom pneumothorax and/or multiple lung cysts are the presenting feature. J. Med. Genet. 47, 281–287 (2010).
Sempau, L., Ruiz, I., Gonzalez-Moran, A., Susanna, X. & Hansen, T. V. New mutation in the Birt–Hogg–Dube gene. Actas Dermosifiliogr. 101, 637–640 (2010).
Boman, P. S., Ousager, L. B., Friis-Hansen, L., Hansen, T. V., Broesby-Olsen, S. & Gerdes, A. M. Is colorectal neoplasia part of the Birt–Hogg–Dube syndrome? J. Gastroenterol. Hepatol. Res. 3, 1039–1042 (2014).
Johannesma, P. C., van den Borne, B. E., Gille, J. J., Nagelkerke, A. F., van Waesberghe, J. T., Paul, M. A. et al. Spontaneous pneumothorax as indicator for Birt–Hogg–Dube syndrome in paediatric patients. BMC Pediatr. 14, 171 (2014).
Kunogi, O. M., Yae, T., Nagashima, O., Hirai, S., Kumasaka, T. & Iwase, A. Pneumothorax developing for the first time in a 73-year-old woman diagnosed with Birt–Hogg–Dube syndrome. Intern. Med. 52, 2453–2455 (2013).
Lim, D. H., Rehal, P. K., Nahorski, M. S., Macdonald, F., Claessens, T., Van, G. M. et al. A new locus-specific database (LSDB) for mutations in the folliculin (FLCN) gene. Hum. Mutat. 31, E1043–E1051 (2010).
Nookala, R. K., Langemeyer, L., Pacitto, A., Ochoa-Montano, B., Donaldson, J. C., Blaszczyk, B. K. et al. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2, 120071 (2012).
Nahorski, M. S., Reiman, A., Lim, D. H., Nookala, R. K., Seabra, L., Lu, X. et al. Birt–Hogg–Dube syndrome-associated FLCN mutations disrupt protein stability. Hum. Mutat. 32, 921–929 (2011).
Hasumi, Y., Baba, M., Ajima, R., Hasumi, H., Valera, V. A., Klein, M. E. et al. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc. Natl Acad. Sci. USA 106, 18722–18727 (2009).
van Steensel, M. A., Verstraeten, V. L., Frank, J., Kelleners-Smeets, N. W., Poblete-Gutierrez, P., Marcus-Soekarman, D. et al. Novel mutations in the BHD gene and absence of loss of heterozygosity in fibrofolliculomas of Birt–Hogg–Dube patients. J. Invest. Dermatol. 127, 588–593 (2007).
Kawasaki, H., Sawamura, D., Nakazawa, H., Hattori, N., Goto, M., Sato-Matsumura, K. C. et al. Detection of 1733insC mutations in an Asian family with Birt–Hogg–Dube syndrome. Br. J. Dermatol. 152, 142–145 (2005).
Lamberti, C., Schweiger, N., Hartschuh, W., Schulz, T., Becker-Wegerich, P., Kuster, W. et al. Birt–Hogg–Dube syndrome: germline mutation in the (C)8 mononucleotide tract of the BHD gene in a German patient. Acta Derm. Venereol. 85, 172–173 (2005).
Acknowledgements
We thank Stine Østergaard and Susanne Smed for excellent laboratory assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Rossing, M., Albrechtsen, A., Skytte, AB. et al. Genetic screening of the FLCN gene identify six novel variants and a Danish founder mutation. J Hum Genet 62, 151–157 (2017). https://doi.org/10.1038/jhg.2016.118
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2016.118
This article is cited by
-
Exons 1–3 deletion in FLCN is associated with increased risk of pneumothorax in Chinese patients with Birt-Hogg-Dubé syndrome
Orphanet Journal of Rare Diseases (2023)
-
Systematic analysis of CNGA3 splice variants identifies different mechanisms of aberrant splicing
Scientific Reports (2023)
-
Splice-site mutation causing partial retention of intron in the FLCN gene in Birt-Hogg-Dubé syndrome: a case report
BMC Medical Genomics (2018)
-
Birt–Hogg–Dubé Syndrome: A Review of Dermatological Manifestations and Other Symptoms
American Journal of Clinical Dermatology (2018)
-
Characterization of a splice-site mutation in the tumor suppressor gene FLCN associated with renal cancer
BMC Medical Genetics (2017)