Abstract
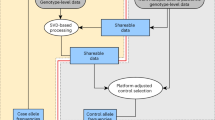
Detecting gene–environment interactions with rare variants is critical in dissecting the etiology of common diseases. Interactions with rare haplotype variants (rHTVs) are of particular interest. At the same time, complex sampling designs, such as stratified random sampling, are becoming increasingly popular for designing case–control studies, especially for recruiting controls. The US Kidney Cancer Study (KCS) is an example, wherein all available cases were included while the controls at each site were randomly selected from the population by frequency matching with cases based on age, sex and race. There is currently no rHTV association method that can account for such a complex sampling design. To fill this gap, we consider logistic Bayesian LASSO (LBL), an existing rHTV approach for case–control data, and show that its model can easily accommodate the complex sampling design. We study two extensions that include stratifying variables either as main effects only or with additional modeling of their interactions with haplotypes. We conduct extensive simulation studies to compare the complex sampling methods with the original LBL methods. We find that, when there is no interaction between haplotype and stratifying variables, both extensions perform well while the original LBL methods lead to inflated type I error rates. However, when such an interaction exists, it is necessary to include the interaction effect in the model to control the type I error rate. Finally, we analyze the KCS data and find a significant interaction between (current) smoking and a specific rHTV in the N-acetyltransferase 2 gene.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Biswas, S. & Lin, S. Logistic Bayesian LASSO for identifying association with rare haplotypes and application to age-related macular degeneration. Biometrics 68, 587–597 (2012).
Biswas, S., Xia, S. & Lin, S. Detecting rare haplotype-environment interaction with logistic Bayesian LASSO. Genet. Epidemiol. 38, 31–41 (2014).
Zhang, Y. & Biswas, S. An improved version of logistic Bayesian LASSO for detecting rare haplotype-environment interactions with application to lung cancer. Cancer Inform. 14, 11–16 (2015).
Zhang, Y., Lin, S. & Biswas, S. Detecting rare haplotype-environment interaction under uncertainty of gene-environment independence assumption. Biometrics 73, 344–355 (2017).
Biswas, S. & Papachristou, C. Evaluation of logistic Bayesian LASSO for identifying association with rare haplotypes. BMC Proc. 8, S54 (2014).
Datta, A. S., Zhang, Y., Zhang, L. & Biswas, S. Association of rare haplotypes on ULK4 and MAP4 genes with hypertension. BMC Proc. 10, 363–369 (2016).
Wang, M. & Lin, S. Detecting associations of rare variants with common diseases: collapsing or haplotyping? Brief Bioinform. 16, 759–768 (2015).
Datta, A. S. & Biswas, S. Comparison of haplotype-based statistical tests for disease association with rare and common variants. Brief Bioinform. 17, 657–671 (2016).
Korn, E. L. & Graubard, B. Analysis of Health Surveys, (Wiley, New York, NY, USA, 1999).
Scott, A. J. & Wild, C. J. Case-control studies with complex sampling. Appl. Stat. 50, 389–401 (2001).
Digaetano, R., Graubard, B., Rao, S., Severynse, J. & Wacholder, S . Sampling racially matched population controls for case-control studies: using DMV lists and oversampling minorities. (2003) https://fcsm.sites.usa.gov/files/2014/05/2003FCSM_DiGaetano.pdf (accessed 8 August 2016).
Colt, J. S., Schwartz, K., Graubard, B. I., Davis, F., Ruterbusch, J., DiGaetano, R. et al. Hypertension and risk of renal cell carcinoma among white and black Americans. Epidemiology 22, 797–804 (2011).
Purdue, M. P., Moore, L. E., Merino, M. J., Boffetta, P., Colt, J. S., Schwartz, K. L. et al. An investigation of risk factors for renal cell carcinoma by histologic subtype in two case-control studies. Int. J. Cancer 132, 2640–2647 (2013).
Hofmann, J. N., Schwartz, K., Chow, W. H., Ruterbusch, J. J., Shuch, B. M., Karami, S. et al. The association between chronic renal failure and renal cell carcinoma may differ between black and white Americans. Cancer Causes Control 24, 167–174 (2013).
Semenza, J. C., Ziogas, A., Largent, J., Peel, D. & Anton-Culver, H. Gene-environment interactions in renal cell carcinoma. Am. J. Epidemiol. 153, 851–859 (2001).
Moore, L. E., Brennan, P., Karami, S., Menashe, I., Berndt, S. I., Dong, L. et al. Apolipoprotein E/C1 locus variants modify renal cell carcinoma risk. Cancer Res. 69, 8001–8008 (2009).
Chow, W., Dong, L. M. & Devesa, S. S. Epidemiology and risk factors for kidney cancer. Nat. Rev. Urol. 7, 245–257 (2010).
Purdue, M. P., Johansson, M., Zelenika, D., Toro, J. R., Scelo, G., Moore, L. E. et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat. Genet. 43, 60–65 (2011).
Li, Y. & Graubard, B. I. Pseudo semiparametric maximum likelihood estimation exploiting gene environment independence for population-based case-control studies with complex samples. Biostatistics 13, 711–723 (2012).
Longuemaux, S., Delomenie, C., Gallou, C., Mejean, A., Vincent-Viry, M., Bouvier, R. et al. Candidate genetic modifiers of individual susceptibility to renal cell carcinoma: a study of polymorphic human xenobiotic-metabolizing enzymes. Cancer Res. 59, 2903–2908 (1999).
Gelman, A., Carlin, J. B., Stern, H. S. & Rubin, D. B. Bayesian Data Analysis, (Chapman and Hall/CRC, Boca Raton, FL, USA, 2003).
Chatterjee, N. & Carroll, R. Semiparametric maximum likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika 92, 399–418 (2005).
Mukherjee, B., Zhang, L., Ghosh, M. & Sinha, S. Semiparametric Bayesian analysis of case-control data under conditional gene-environment independence. Biometrics 63, 834–844 (2007).
Weir, B. S. Genetic Data Analysis II, (Sinauer Associates Inc, Sunderland, MA, USA, 1996).
Mukherjee, B. & Chatterjee, N. Exploiting gene-environment independence for analysis of case-control studies: An empirical-Bayes type shrinkage estimator to trade off between bias and efficiency. Biometrics 64, 685–694 (2008).
Prentice, R. L. & Pyke, R. Logistic disease incidence models and case-control studies. Biometrika 66, 403–411 (1979).
Kwee, L. C., Epstein, M. P., Manatunga, A. K., Duncan, R., Allen, A. S. & Satten, G. A. Simple methods for assessing haplotype-environment interactions in case-only and case-control studies. Genet. Epidemiol. 31, 75–90 (2007).
Lake, S. L., Lyon, H., Tantisira, K., Silverman, E. K., Weiss, S. T., Laird, N. M. et al. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum. Hered. 55, 56–65 (2003).
Deitz, A. C., Rothman, N., Rebbeck, T. R., Hayes, R. B., Chow, W. H., Zheng, W. et al. Impact of misclassification in genotype-exposure interaction studies: example of N-Acetyltransferase 2 (NAT2), smoking, and bladder cancer. Cancer Epidemiol. Biomarkers Prev. 13, 1543–1546 (2004).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Burkett, K., Graham, J. & McNeney, B. hapassoc: software for likelihood inference of trait associations with SNP haplotypes and other attributes. J. Stat. Softw. 16, 1–19 (2006).
Scott, A. J. & Wild, C. J . in Analysis of Survey Data (eds Chambers, R. L., Skinner, C. J.) 109–120 (Wiley, Chichester, England, 2003).
Xia, S. & Lin, S. Detecting longitudinal effects of haplotypes and smoking on hypertension using B-Splines and Bayesian LASSO. BMC Proc. 8, S85 (2014).
Wang, M. & Lin, S. FamLBL: detecting rare haplotype disease association based on common SNPs using case-parent triads. Bioinformatics 30, 2611–2618 (2014).
Acknowledgements
This work was partially supported by the grant R03CA171011 from the National Cancer Institute, NIH and by allocations of computing times from the Texas Advanced Computing Center at the University of Texas at Austin. The US Kidney Cancer Study was supported by the Intramural Research Program of the NIH, National Cancer Institute. We are thankful to the two anonymous referees for their constructive comments and suggestions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, Y., Hofmann, J., Purdue, M. et al. Logistic Bayesian LASSO for genetic association analysis of data from complex sampling designs. J Hum Genet 62, 819–829 (2017). https://doi.org/10.1038/jhg.2017.43
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2017.43
This article is cited by
-
Development and validation of a novel nomogram model for predicting delayed graft function in deceased donor kidney transplantation based on pre-transplant biopsies
BMC Nephrology (2024)
-
Bayesian variable selection for high-dimensional data with an ordinal response: identifying genes associated with prognostic risk group in acute myeloid leukemia
BMC Bioinformatics (2021)