Abstract
DNA methylation in gene promoters causes gene silencing and is a common event in cancer development and progression. The ability of aberrant methylation events to serve as diagnostic and prognostic markers is being appreciated for many cancers, including prostate cancer. Using quantitative MethyLight technology, we evaluated the relationship between HOXD3 methylation and clinicopathological parameters including biochemical recurrence, pathological stage, Gleason score (GS), and Gleason pattern in a series of 232 radical prostatectomies performed between 1998 and 2001. HOXD3 methylation was significantly greater in GS 7 cancers vs GS≤6 cancers (P-value <0.001) as well as pT3/pT4 vs pT2 cancers (P-value <0.001). The proportion of cases with high methylation in GS 7 vs ≤GS 6 and pT3/pT4 vs pT2 were also significantly different (P-values=0.002 and 0.005, respectively). There were also significant increases in methylation from Gleason pattern 2–3 and from pattern 3 to 4/5 (paired t-test P-values=0.01 and <0.001, respectively), whereas methylation from lymph node metastases was decreased when compared with matched tumor tissue (P-value=0.029). HOXD3 methylation was associated with biochemical recurrence in univariate analysis (P-value=0.043) and showed evidence for interaction with pathological stage as a predictor variable in Cox regression analysis (P-value=0.028). The results indicate that HOXD3 methylation distinguishes low-grade prostate cancers from intermediate and high-grade ones and may also have prognostic value when considered together with pathological stage.
Similar content being viewed by others
Main
Prostate cancer (PCa) is the most common malignancy for North American men, and in 2009 it is estimated that it will account for >27 000 deaths in the United States.1 The widespread acceptance of routine PSA testing has led to dramatic increases in PCa diagnoses over the past two decades, while the benefits of such testing remain controversial.2, 3 Therefore, predicting disease course and outcome of diagnosed cases has received attention recently as many of these cancers are of a slow growing or indolent form, whereas others may progress rapidly and result in metastasis and death. The best prognostic indicator for PCa, Gleason score (GS), characterizes the glandular architecture of the prostate and assigns a score based on ‘de-differentiation’ of the carcinoma.4 The extent of de-differentiation is represented by Gleason patterns, which range from 1 to 5. Pattern 3 and pattern 4 represent an important transition from low- to high-grade carcinoma, and can influence patient prognosis depending on the relative proportions present in the cancer. Pure pattern 3 (GS 6) PCas are low grade, whereas pure pattern 4 (GS 8) cancers are high grade.5 GS 7 cancers (composed of glandular patterns 3 and 4 in variable amounts) are considered intermediate grade. Pretreatment identification of GS 7 cancers (ie presence of Gleason pattern 4) is considered key to distinguish indolent or clinically insignificant PCa from those that require treatment.
DNA methylation is a well-characterized epigenetic event involving the addition of a methyl group to cytosine bases that precede a guanine (CpG). This modification has an important function in promoting chromosomal stability and regulating gene expression.6, 7 In mammals, enriched DNA stretches of CpG dinucleoties known as CpG islands are common in the promoter regions of ∼50% of genes.8 With the exception of tissue-specific methylation patterns, most promoter CpG islands are hypomethylated, whereas ∼70% of total CpG dinucleotides exist in a methylated state.9 Aberrant methylation patterns occur frequently in cancers, including an overall hypomethylation of genomic CpGs.10 Gene-specific anomalies are also common, however, and can lead to the inactivation of tumor suppressor genes or to the activation of oncogenes.8, 11 The former arises through hypermethylation of promoters, whereas the latter occurs when promoter CpG islands become hypomethylated. Importantly, promoter methylation events are being recognized for their potential as both diagnostic and prognostic indicators for a variety of malignancies including bladder, prostate, and colon cancers.12, 13
We have previously identified HOXD3 promoter hypermethylation in PCa through a genome-wide CpG island microarray screen of GS 6 and GS 8 cases.14 To assess the contribution of HOXD3 methylation as a diagnostic/progression marker in this study, we have now examined a larger series of PCa using a quantitative methylation approach, and we analyzed the relationship of HOXD3 methylation levels with clinicopathological parameters.
MATERIALS AND METHODS
Patients and Pathology
A total of 232 patients diagnosed with PCa between 1998 and 2001 at the University Health Network (UHN) in Toronto were included in this study. We included an additional 11 PCa cases (for a total of 243) with lymph node metastases to attain a total of 16 cases in this subgroup. Both the primary cancer and the metastatic lymph node (MLN) lesion from these cases were analyzed. These cases were not included in the survival analysis, as there was an insufficient time span for follow-up data to be collected (diagnosed between 2002 and 2009). Patient consent was obtained for accrual of removed tissue following radical prostatectomy into the UHN tissue bank. All samples and clinical and pathological follow-up information were obtained according to the protocols approved by the Research Ethics Board at Mount Sinai Hospital, Toronto and UHN, Toronto. All patients who received neo-adjuvant therapy before radical prostatectomy were excluded from the study.
The complete set of hematoxylin and eosin (H&E)-stained slides from each prostatectomy was collected and reviewed by an expert pathologist (TVDK) to confirm GS (WHO/ISUP criteria), stage (TNM), and surgical margin status. For each case, a subset of slides was selected based on the presence of carcinoma with specific Gleason patterns representing the overall GS. Both a pattern 3 and a pattern 4 were selected for Gleason 7 cases where possible. Tumor areas representative of each Gleason pattern were marked on the H&E-stained slides corresponding to an area of at least 80% neoplastic cellularity. Matched normal tissue containing at least 50% glandular content was also selected for each case where possible.
DNA Extraction
Formalin-fixed, paraffin-embedded tissue blocks matching the selected H&E slides were sectioned at a thickness of 10 μm. These tissue slides were then superimposed on H&E slides and each area of cancer was outlined. The circled areas of tissue slides were then scraped with a scalpel and tissue placed into 1.5 ml tubes.
DNA was extracted from tissues using QIAamp DNA Mini Kit (Qiagen, Mississauga, ON, Canada) with a modified protocol. Briefly, 270 μl of buffer ATL was added to tissue followed by 30 μl of proteinase K. The tissue was digested overnight at 56°C and 20 μl of proteinase K was added the following day. The tissue was then incubated at 56°C further for 1 h, and an equal volume (320 μl) of buffer AL was added with an incubation of 70°C for 10 min. One volume of ethanol (320 μl) was then added, with the remainder of the steps performed according to the manufacturer's recommended protocol for tissue DNA extraction.
Sodium Bisulfite Modification and MethyLight
A measure of 400 ng of extracted DNA was converted using the EZ DNA Methylation Gold Kit (Zymo Research, Oragne, CA, USA) according to the manufacturer's protocol and eluted to a final concentration of 20 ng/μl.
The quantitative MethyLight assay was performed as described earlier using 20 ng converted DNA.15 The reactions were carried out in a volume of 30 μl in 96-well plates on an ABI 7500 Real-Time PCR system. Standard curves were generated using serial dilutions of positive control supermethylated DNA for the gene of interest and Alu repeats. Percentage of methylated reference (PMR) for a gene was calculated using Alu repeats as reference as follows: (HOXD3/Alu fluorescence quantity ratio for modified specimen DNA)/(HOXD3/Alu ratio for supermethylated DNA) × 100%. For each case, at least one or more (up to six) tumor foci were analyzed. All foci were analyzed in duplicate. Primer/probe sequences for the HOXD3-assayed region were as follows: (Forward) 5′-TTA AAG GTT TAT GGT TGC GC-3′; (Reverse) 5′-TTA CGA ACA CTA AAC TAC ACC CG-3′; (Probe) 5′FAM-ACA AAA CGT TCC CGA CGC TTC TAA AA-BHQ1-3′.
Statistical Analysis
PMR scores for each sample analyzed were obtained from averaging duplicate runs. When multiple foci were analyzed for each PCa specimen, an individual PMR value was assigned to each case based on an average of the PMR values obtained for each focus within that case.
Univariate disease-free survival (biochemical recurrence) was calculated using the log-rank test and Kaplan–Meier method. Multivariate Cox proportional hazards regression analysis was used to analyze individual contributions of each variable to disease-free survival. We used both a Wald method and likelihood ratio test (LRT) to analyze the data. The LRT tests for significance of variables by comparing the full model to a reduced model, with the reduced model missing the variable that is being tested for significance. For both univariate and multivariate analyses, HOXD3 methylation was separated into two groups of high methylation (HM) and low methylation (LM) based on a third quartile threshold as described earlier.16, 17 Briefly, the data for 232 cases was split based on PMR values above the third quartile vs those in the lower three quartiles. Those PMRs in the upper quartile were classified as HM, whereas those below this value were considered LM.
Analysis of the relationship between average HOXD3 methylation and GS, pattern and stage was done using the Mann–Whitney U-test. Pearson χ2 tests were used to analyze proportional differences between HM cases in each category of GS, pattern, and stage. The Fisher exact method was used to replace the Pearson tests when spreadsheet cell counts were <5. Further, we performed paired t-tests for matched Gleason pattern analysis and matched MLN/primary cancer analysis. For Gleason pattern data, we used the PMR from each of the cancer foci in every case to establish HM and LM categories, again separating based on the third quartile.
For all described methods, P-values of ≤0.05 were considered significant. Statistics were performed using SPSS (Chicago, IL, USA) and R statistical software.
RESULTS
Clinical and Pathological Variables
Table 1 displays the clinicopathological characteristics of the 232 patients included in the study. The average age of patients was 61 years. A total of 85 patients (36.5%) had biochemical recurrence, whereas the mean follow-up time was 1600 days (range 63–3460).
Methylation in Normal Tissue, Primary Cancer, and Metastasis
We initially analyzed average methylation values in PCa specimens (519 total from 232 patients) compared with matched normal tissue (229 total specimens). The average PMR for PCa was significantly higher than normal tissue (27.4 vs 7.6, respectively, Mann–Whitney P-value <0.001). χ2 analysis also showed a greater proportion of HM cases in cancer specimens compared with normal (26.4 vs 0.4%, P-value <0.001). Furthermore, we performed receiver–operator curve analysis to determine the suitability of this marker to accurately distinguish cancer from normal tissues. We chose the PMR value that maximized sensitivity and specificity in identifying cancer vs normal tissue. The area under the curve was 0.798 (95% confidence interval: 0.767–0.830), with a sensitivity and specificity of 61.8 and 86.9%, respectively (Supplementary Figure 1).
We also analyzed PCa that had metastasized to lymph nodes (MLN, Tables 2 and 4) and the corresponding primary PCa using a paired t-test. Interestingly, we found significant decreases when comparing matched MLN tissue to the primary tumor (average PMR difference −16.9, P-value=0.029).
HOXD3 Methylation and GS/Pathological Stage
Next, we determined whether methylation of HOXD3 increased in conjunction with increasing GS. The data were separated into groups of low (GS≤6), intermediate (GS=7), and high grade (GS≥8). Average PMR values for these groups are given in Table 2. Using an ANOVA model, we found a significant difference (P-value <0.001) between GS≤6, 7, and ≥8 PCa. Further subdivision of the categories revealed increases of average PMR between the GS 7 and ≤6 subgroups (28.9 vs 18.8, respectively; P-value <0.001) and a higher proportion of HM cases in the GS 7 group vs the GS≤6 group (30.8 vs 12.9%, P-value=0.002). We also tested for differences between Gleason 7 cases that were predominant pattern 3 (3+4) vs predominant pattern 4 (4+3). Although a comparison of PMR averages revealed greater methylation in 4+3 vs 3+4 approaching significance (33.5 vs 27.0, respectively; P-value=0.076), the number of HM cases in each group was similar (χ2 P-value=0.605).
Similarly, we examined the relationship of HOXD3 methylation with pathological stage (Table 3). Again, we found significant differences overall between organ confined pT2 cases and invasive pT3/pT4 cases, with greater average methylation (32.3 vs 21.7, P-value <0.001) and a greater proportion of HM cases (35.2 vs 18.8%, P-value=0.005) in pT3/pT4 cases vs the pT2 group.
HOXD3 Methylation and Gleason Pattern
Next, we analyzed methylation differences between each Gleason pattern. Patterns 4 and 5 were grouped into the same high-grade category due to a small number of observations for pattern 5 carcinoma (n=8). Initial ANOVA analysis demonstrated significant differences between all groups (P-value <0.001), whereas comparison of means between subdivided groups showed significant stepwise PMR increases comparing pattern 2 to 3 (6.8 vs 23.2, respectively; P-value <0.001) and pattern 3 to 4/5 (23.2 vs 35.3, respectively; P-value <0.001). Furthermore, we compared the methylation levels from identical Gleason patterns obtained from different GS. Therefore, pattern 3 from Gleason 7 was compared with pattern 3 from Gleason 6, and pattern 4 from Gleason 8 was compared with pattern 4 from Gleason 7. Although we did not observe significant differences for average PMR or HM frequencies in either of the subdivided categories, there was a greater PMR average of pattern 3 from GS 7 vs pattern 3 from GS≤6 that approached significance (24.9 vs 20.9, respectively; Mann–Whitney and χ2 P-values=0.07 and 0.190, respectively).
In addition, we used a paired t-test to distinguish whether HOXD3 methylation continues to be associated with higher Gleason patterns within individual cases (Table 4). We detected increases in methylation between matched normal tissue and each of patterns 3 and 4/5. Pairwise comparisons of Gleason pattern 3 vs 2 (average PMR difference 10.5, P-value=0.01) and 4/5 vs 3 (average PMR difference 9.3, P-value <0.001) also showed significant increases in higher Gleason patterns.
Biochemical Progression-Free Survival
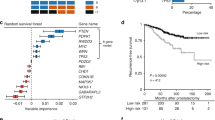
Univariate log-rank analysis of GS, stage, and surgical margin status showed that each are significant predictors of biochemical recurrence (Figure 1a–c), indicating that the series of patients included in this study is representative of populations studied earlier.18, 19
We next examined the relationship between biochemical recurrence and the HM or LM groups. Univariate analysis of those in the HM group vs LM group revealed significant differences in disease-free survival (Figure 1d, log-rank P-value=0.043). The probability of biochemical progression in the HM group was 43.1%, whereas in the LM group it was 34.5%. Furthermore, we performed multivariate Cox regression analysis of our data (Table 5). Significant predictors of biochemical progression-free survival included GS, pathological stage, and surgical margin status, whereas age and HOXD3 methylation status were not significant predictors. We included interaction terms into the regression analysis to determine whether there were any associations between HOXD3 methylation and pathological variables with respect to progression-free survival. We found that the interaction involving pathological stage and HOXD3 methylation was a significant predictor of biochemical recurrence, whereas the remaining interactions of HOXD3 methylation with GS, age, and surgical margins were not. Thus, HOXD3 methylation in combination with known pathological stage (invasive stages in particular) is an independent predictor of biochemical recurrence.
DISCUSSION
Homeobox genes are a family of transcription factors that were first identified in Drosophila and characterized by their ability to cause large-scale patterning abnormalities when mutated.20, 21 These genes contain a highly conserved 183 bp region encoding a 61 amino acid helix-turn-helix DNA-binding motif known as the homeodomain.22, 23 In humans, there are over 200 such genes containing this motif, and many of them have been identified as aberrantly expressed in cancers.24, 25 For example, overexpression of HOXC6, PAX2, and BP1 have been described in PCa.26, 27, 28 We have previously shown that methylation of homeobox genes is a common event in PCa,14 whereas others have shown similar results in so-called methylation ‘hot spots’ within HOX loci.29, 30 In particular, we demonstrated methylation of the HOXD3 region in primary PCa and that this methylation leads to gene silencing in DU-145 cells.14
Our results show that HOXD3 methylation is elevated in intermediate grade GS 7 compared with low-grade GS 6 tumors. Gleason 6–7 represents a biologically and clinically important switch point as GS 7 cancers have an unfavorable prognosis compared with those of lower grades. Assigning a HOXD3 methylation value in conjunction with GS may have important clinical applications. GSs assigned on a biopsy are often inaccurate as compared with the GS assigned in the corresponding prostatectomy specimen and may lead to inappropriate treatment strategies.31, 32 Similarly, clinical staging lacks accuracy as many cancers seem to be understaged if compared with the pathological stage. Therefore, a marker that is indicative of a higher grade and stage of PCa could be helpful in clinical decision making. HOXD3 methylation may also be detectable in voided urine samples or following prostatic massage as has been shown for other methylation markers,33 thus allowing a diagnosis/prognosis before any further invasive exams as detection of methylation would indicate the presence of high-grade disease.
A unique aspect of this study was the large-scale comparison of different Gleason patterns from matched PCa specimens. We were able to show that HOXD3 methylation is significantly greater in high-grade vs low-grade Gleason patterns, either when comparing overall averages or when matching patterns to the same patient. Importantly, we observed increases in PMR for HOXD3 progressing from matched normal to pattern 3 and further onto high-grade patterns 4 or 5. It is possible that methylation of HOXD3, along with other genetic and/or epigenetic events, is responsible for progressive de-differentiation of PCa foci. For example, loss of expression of the homeobox gene PDX1 has been shown to have an inverse correlation with increasing Gleason pattern.34 In addition, loss of the androgen responsive gene NKX3.1 (required for differentiating prostate epithelial cells) has been reported as HOXC8 overexpression contributing to a loss of differentiation.35, 36, 37 Hence, it is likely that the proper expression of specific homeobox genes (including HOXD3) is crucial for the maintenance of a differentiated tissue phenotype, which is concordant with the role that homeobox genes have in the development of tissue-specific architecture.38, 39 However, an alternative to this would be that HOXD3 methylation may be an epiphenomenon of these processes instead of a cause. Future work is necessary to address this issue.
Interestingly, we did not observe any overall differences in methylation between normal tissue and Gleason pattern 2 carcinomas. In fact, paired analysis of matched normal with pattern 2 revealed a significant decrease in methylation. One possible explanation may be that pattern 2 carcinomas often arise from the transition zone of the prostate, and transition zone carcinomas are typically not as aggressive as cancers in the peripheral zone.40, 41 DNA methylation patterns may differ according to different prostatic zones from which the cancers arise, and thus may relate to good or poor prognosis. We also found that lymph node metastatic tissue had less HOXD3 methylation than corresponding matched primary PCa. A similar trend of increased methylation in high-grade patterns 4 or 5 with reduced methylation in MLN and bone tissue for the ER-β has previously been reported in PCa.42 It is possible that PCa cells that migrate into a different niche develop a unique epigenetic identity due to different environmental factors and/or selection pressures. Alternatively, HOXD3 methylation might confer a growth advantage in primary PCa, whereas a subset of cells lacking HOXD3 methylation in the primary tumor may be more capable of metastasizing. Further work is also needed to clarify this issue.
On multivariate Cox regression analysis with classical prognostic markers (GS, stage, surgical margin status), the contribution of methylation status to the model was not statistically significant. There may be multiple reasons for this occurrence. First, our cohort was also limited by relatively few Gleason ≥8 cases (n=24), which may have affected the recurrence analysis. Second, methylation of this locus might be one of a panel of methylation markers, which together predict PCa disease course with a high degree of accuracy. Prior studies have shown the usefulness of multigene methylation panels for diagnosis and prognosis of PCa.16, 43 It is worthwhile to note, however, that we observed an interaction between HOXD3 methylation and pathological stage in predicting biochemical recurrence, indicating that HOXD3 HM maybe useful for determining which higher stage cancers will likely recur. Thus, HOXD3 may also be clinically useful after surgery when considered along with stage, perhaps warranting more aggressive treatment in patients with pT3a tumors.
The role that HOXD3 may have in PCa development and progression has not been elucidated. Similar to PCa, this gene has been shown to be methylated in A549 lung cancer cells.29 Functional analysis of HOXD3 has shown that overexpression drives angiogenesis, and in lung cancer A549 cells it contributes to a metastatic phenotype via coordinate expression of metastasis-related genes and through TGFβ dependent and independent mechanisms.44, 45 Whether the paradoxical situation of methylation and increased expression leading to metastasis exists in PCa is not known. Our MLN methylation results suggest, however, that methylation may be lost in a subset of cases, which could lead to a metastatic phenotype. It may be that complex mechanisms exist to maintain proper expression of HOXD3 in tissue-specific settings, and DNA methylation may only partially contribute.
In conclusion, we have characterized HOXD3 promoter methylation in a series of PCa cases and analyzed its relationship to classical clinicopathological parameters. We have shown that methylation levels are significantly greater in intermediate-grade vs low-grade GSs, and have shown increased methylation with loss of tumor differentiation according to Gleason patterns. Future work is necessary to elucidate both the clinical utility and functional relevance of HOXD3, including possible detection in urine/serum of PCa patients as well as the role of methylation and/or abnormal expression of this gene in PCa progression.
References
Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225–249.
Andriole GL, Crawford ED, Grubb III RL, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med 2009;360:1310–1319.
Schroder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med 2009;360:1320–1328.
Gleason DF . Classification of prostatic carcinomas. Cancer Chemother Rep 1966;50:125–128.
Lopez-Beltran A, Mikuz G, Luque RJ, et al. Current practice of Gleason grading of prostate carcinoma. Virchows Arch 2006;448: 111–118.
Eden A, Gaudet F, Waghmare A, et al. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003;300:455.
Jones PA, Baylin SB . The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–428.
Baylin SB . DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2005;2 (Suppl 1):S4–S11.
Brena RM, Costello JF . Genome-epigenome interactions in cancer. Hum Mol Genet 2007;16 Spec No 1:R96–R105.
Wilson AS, Power BE, Molloy PL . DNA hypomethylation and human diseases. Biochim Biophys Acta 2007;1775:138–162.
Lee PS, Teaberry VS, Bland AE, et al. Elevated MAL expression is accompanied by promoter hypomethylation and platinum resistance in epithelial ovarian cancer. Int J Cancer 2009;126:1378–1389.
Laird PW . The power and the promise of DNA methylation markers. Nat Rev Cancer 2003;3:253–266.
Kim YK, Kim WJ . Epigenetic markers as promising prognosticators for bladder cancer. Int J Urol 2009;16:17–22.
Kron K, Pethe V, Briollais L, et al. Discovery of novel hypermethylated genes in prostate cancer using genomic CpG island microarrays. PLoS One 2009;4:e4830.
Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:E32.
Rosenbaum E, Hoque MO, Cohen Y, et al. Promoter hypermethylation as an independent prognostic factor for relapse in patients with prostate cancer following radical prostatectomy. Clin Cancer Res 2005;11:8321–8325.
Henrique R, Ribeiro FR, Fonseca D, et al. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin Cancer Res 2007;13:6122–6129.
Weiss G, Cottrell S, Distler J, et al. DNA methylation of the PITX2 gene promoter region is a strong independent prognostic marker of biochemical recurrence in patients with prostate cancer after radical prostatectomy. J Urol 2009;181:1678–1685.
Daneshmand S, Quek ML, Lin E, et al. Glucose-regulated protein GRP78 is up-regulated in prostate cancer and correlates with recurrence and survival. Hum Pathol 2007;38:1547–1552.
Lewis EB . A gene complex controlling segmentation in Drosophila. Nature 1978;276:565–570.
Scott MP, Weiner AJ . Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci USA 1984;81:4115–4119.
Gehring WJ . Exploring the homeobox. Gene 1993;135:215–221.
Gehring WJ, Affolter M, Burglin T . Homeodomain proteins. Annu Rev Biochem 1994;63:487–526.
Christensen KL, Patrick AN, McCoy EL, et al. The six family of homeobox genes in development and cancer. Adv Cancer Res 2008;101:93–126.
Samuel S, Naora H . Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer 2005;41:2428–2437.
Miller GJ, Miller HL, van Bokhoven A, et al. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res 2003;63:5879–5888.
Khoubehi B, Kessling AM, Adshead JM, et al. Expression of the developmental and oncogenic PAX2 gene in human prostate cancer. J Urol 2001;165 (6 Pt 1):2115–2120.
Schwartz AM, Man YG, Rezaei MK, et al. BP1, a homeoprotein, is significantly expressed in prostate adenocarcinoma and is concordant with prostatic intraepithelial neoplasia. Mod Pathol 2009;22:1–6.
Rauch T, Li H, Wu X, et al. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res 2006;66:7939–7947.
Rauch T, Wang Z, Zhang X, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA 2007;104:5527–5532.
Koksal IT, Ozcan F, Kadioglu TC, et al. Discrepancy between Gleason scores of biopsy and radical prostatectomy specimens. Eur Urol 2000;37:670–674.
Pinthus JH, Witkos M, Fleshner NE, et al. Prostate cancers scored as Gleason 6 on prostate biopsy are frequently Gleason 7 tumors at radical prostatectomy: implication on outcome. J Urol 2006;176:979–984; discussion 984.
Goessl C, Muller M, Heicappell R, et al. DNA-based detection of prostate cancer in urine after prostatic massage. Urology 2001;58: 335–338.
Jonmarker S, Glaessgen A, Culp WD, et al. Expression of PDX-1 in prostate cancer, prostatic intraepithelial neoplasia and benign prostatic tissue. APMIS 2008;116:491–498.
He WW, Sciavolino PJ, Wing J, et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics 1997;43:69–77.
Asatiani E, Huang WX, Wang A, et al. Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res 2005;65:1164–1173.
Waltregny D, Alami Y, Clausse N, et al. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate 2002;50:162–169.
Abate-Shen C, Shen MM, Gelmann E . Integrating differentiation and cancer: the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation 2008;76:717–727.
Duverger O, Morasso MI . Role of homeobox genes in the patterning, specification, and differentiation of ectodermal appendages in mammals. J Cell Physiol 2008;216:337–346.
Noguchi M, Stamey TA, Neal JE, et al. An analysis of 148 consecutive transition zone cancers: clinical and histological characteristics. J Urol 2000;163:1751–1755.
Shannon BA, McNeal JE, Cohen RJ . Transition zone carcinoma of the prostate gland: a common indolent tumour type that occasionally manifests aggressive behaviour. Pathology 2003;35:467–471.
Zhu X, Leav I, Leung YK, et al. Dynamic regulation of estrogen receptor-beta expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol 2004;164:2003–2012.
Richiardi L, Fiano V, Vizzini L, et al. Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. J Clin Oncol 2009;27:3161–3168.
Hamada J, Omatsu T, Okada F, et al. Overexpression of homeobox gene HOXD3 induces coordinate expression of metastasis-related genes in human lung cancer cells. Int J Cancer 2001;93:516–525.
Miyazaki YJ, Hamada J, Tada M, et al. HOXD3 enhances motility and invasiveness through the TGF-beta-dependent and -independent pathways in A549 cells. Oncogene 2002;21:798–808.
Acknowledgements
This work was supported by the National Cancer Institute of Canada/Canadian Prostate Cancer Research Initiative #18568 (BB, TVDK, NF, LB) and the Ontario Student Opportunity Trust Fund (KK, LL).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
Supplementary information
Rights and permissions
About this article
Cite this article
Kron, K., Liu, L., Pethe, V. et al. DNA methylation of HOXD3 as a marker of prostate cancer progression. Lab Invest 90, 1060–1067 (2010). https://doi.org/10.1038/labinvest.2010.57
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/labinvest.2010.57
Keywords
This article is cited by
-
Exploring targets of TET2-mediated methylation reprogramming as potential discriminators of prostate cancer progression
Clinical Epigenetics (2019)
-
A urine-based DNA methylation assay, ProCUrE, to identify clinically significant prostate cancer
Clinical Epigenetics (2018)
-
Novel Multiplex MethyLight Protocol for Detection of DNA Methylation in Patient Tissues and Bodily Fluids
Scientific Reports (2014)
-
Methylation of PITX2, HOXD3, RASSF1 and TDRD1 predicts biochemical recurrence in high-risk prostate cancer
Journal of Cancer Research and Clinical Oncology (2014)
-
Methylation markers for prostate cancer prognosis: a systematic review
Cancer Causes & Control (2013)