Abstract
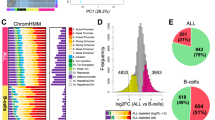
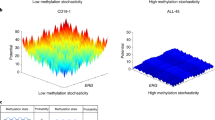
Patients with Down syndrome (DS) and acute lymphoblastic leukemia (ALL) have distinct clinical and biological features. Whereas most DS-ALL cases lack the sentinel cytogenetic lesions that guide risk assignment in childhood ALL, JAK2 mutations and CRLF2 overexpression are highly enriched. To further characterize the unique biology of DS-ALL, we performed genome-wide profiling of 58 DS-ALL and 68 non-DS (NDS) ALL cases by DNA copy number, loss of heterozygosity, gene expression and methylation analyses. We report a novel deletion within the 6p22 histone gene cluster as significantly more frequent in DS-ALL, occurring in 11 DS (22%) and only 2 NDS cases (3.1%) (Fisher's exact P=0.002). Homozygous deletions yielded significantly lower histone expression levels, and were associated with higher methylation levels, distinct spatial localization of methylated promoters and enrichment of highly methylated genes for specific pathways and transcription factor-binding motifs. Gene expression profiling demonstrated heterogeneity of DS-ALL cases overall, with supervised analysis defining a 45-transcript signature associated with CRLF2 overexpression. Further characterization of pathways associated with histone deletions may identify opportunities for novel targeted interventions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Whitlock JA . Down syndrome and acute lymphoblastic leukaemia. Br J Haematol 2006; 135: 595–602.
Forestier E, Izraeli S, Beverloo B, Haas O, Pession A, Michalova K et al. Cytogenetic features of acute lymphoblastic and myeloid leukemias in pediatric patients with Down syndrome: an iBFM-SG study. Blood 2008; 111: 1575–1583.
Maloney KW, Carroll WL, Carroll AJ, Devidas M, Borowitz MJ, Martin PL et al. Down syndrome childhood acute lymphoblastic leukemia has a unique spectrum of sentinel cytogenetic lesions that influences treatment outcome: a report from the Children's Oncology Group. Blood 2010; 116: 1045–1050.
Bercovich D, Ganmore I, Scott LM, wainreb G, Birger Y, Elimelech A et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet 2008; 372: 1484–1492.
Kearney L, Gonzalez De CD, Yeung J, Procter J, Horsley SW, Eguchi-Ishimae M et al. Specific JAK2 mutation (JAK2R683) and multiple gene deletions in Down syndrome acute lymphoblastic leukemia. Blood 2009; 113: 646–648.
Gaikwad A, Rye CL, Devidas M, Heerema NA, Carroll AJ, Izraeli S et al. Prevalence and clinical correlates of JAK2 mutations in Down syndrome acute lymphoblastic leukaemia. Br J Haematol 2009; 144: 930–932.
Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA 2009; 106: 9414–9418.
Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood 2009; 114: 2688–2698.
Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet 2009; 41: 1243–1246.
Hertzberg L, Vendramini E, Ganmore I, Cazzaniga G, Schmitz M, Chalker J et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood 2010; 115: 1006–1017.
Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood 2010; 115: 5312–5321.
Yoda A, Yoda Y, Chiaretti S, Bar-Natan M, Mani K, Rodig SJ et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA 2010; 107: 252–257.
Cario G, Zimmermann M, Romey R, Gesk S, Vater I, Harbott J et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood 2010; 115: 5393–5397.
Ensor HM, Schwab C, Russell LJ, Richards SM, Morrison H, Masic D et al. Demographic, clinical and outcome features of children with acute lymphoblastic leukemia and CRLF2 deregulation: results from the MRC ALL97 clinical trial. Blood 2011; 117: 2129–2136.
Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL . Whole-genome genotyping with the single-base extension assay. Nat Methods 2006; 3: 31–33.
Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res 2006; 16: 1136–1148.
Rozen S, Skaletsky HJ . Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press: Totowa, NJ, 2000, pp 365–386.
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321: 1807–1812.
Scott DA, Klaassens M, Holder AM, Lally KP, Fernandes CJ, Galjaard RJ et al. Genome-wide oligonucleotide-based array comparative genome hybridization analysis of non-isolated congenital diaphragmatic hernia. Hum Mol Genet 2007; 16: 424–430.
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007; 446: 758–764.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res 2008; 36 (Database issue): D480–D484.
Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K et al. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature 2005; 434: 338–345.
Li C, Wong W . Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2001; 2: 0032.1–0032.11.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273.
Peto R, Pike MC, Armitage P, Breslow NE, Cox DR, Howard SV et al. Design and analysis of randomized clinical trials requiring prolonged observation of each patient. II. analysis and examples. Br J Cancer 1977; 35: 1–39.
Feuk L, Carson AR, Scherer SW . Structural variation in the human genome. Nat Rev Genet 2006; 7: 85–97.
Zhang F, Carvalho CM, Lupski JR . Complex human chromosomal and genomic rearrangements. Trends Genet 2009; 25: 298–307.
Marzluff WF, Wagner EJ, Duronio RJ . Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 2008; 9: 843–854.
Ramensky V, Bork P, Sunyaev S . Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002; 30: 3894–3900.
Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 2006; 43: 295–305.
Mathe E, Olivier M, Kato S, Ishioka C, Hainaut P, Tavtigian SV . Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res 2006; 34: 1317–1325.
Chen J, Odenike O, Rowley JD . Leukaemogenesis: more than mutant genes. Nat Rev Cancer 2010; 10: 23–36.
Ellis L, Atadja PW, Johnstone RW . Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther 2009; 8: 1409–1420.
Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ . The human and mouse replication-dependent histone genes. Genomics 2002; 80: 487–498.
Berger SL . The complex language of chromatin regulation during transcription. Nature 2007; 447: 407–412.
Greaves MF, Wiemels J . Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer 2003; 3: 639–649.
Acknowledgements
We thank Susan Hilsenbeck, Jian Wang, Alexander Yu and Charlotte Ahern for biostatistical support; Daryl Scott for assistance with genomic quantitative PCR; Brandon Ballard for assistance with PCR and data analysis; Emanuela Giarin for assistance with AIEOP sample annotation; Richard Harvey for discussion and sharing preliminary bioinformatic analysis; and the patients and families whose participation made this work possible. This work was supported in part by a Bear Necessities Pediatric Research Foundation Grant; Children's Cancer Research Foundation Grant; National Institutes of Health Pediatric Oncology Clinical Research Training Grant CA90433-06; Gillson Longenbaugh Foundation; Kurt Groten Family Research Scholars' Program (KRR); and the National Cancer Institute (NCI; SPEC U01 CA114762 and Genomics Core of the NYU Cancer Institute P30 CA016087, WLC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Loudin, M., Wang, J., Eastwood Leung, HC. et al. Genomic profiling in Down syndrome acute lymphoblastic leukemia identifies histone gene deletions associated with altered methylation profiles. Leukemia 25, 1555–1563 (2011). https://doi.org/10.1038/leu.2011.128
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/leu.2011.128
Keywords
This article is cited by
-
Learning biologically-interpretable latent representations for gene expression data
Machine Learning (2023)
-
Identify Down syndrome transcriptome associations using integrative analysis of microarray database and correlation-interaction network
Human Genomics (2018)
-
Micro RNAs and DNA methylation are regulatory players in human cells with altered X chromosome to autosome balance
Scientific Reports (2017)
-
The biology, pathogenesis and clinical aspects of acute lymphoblastic leukemia in children with Down syndrome
Leukemia (2016)
-
Clinical and genetic features of pediatric acute lymphoblastic leukemia in Down syndrome in the Nordic countries
Journal of Hematology & Oncology (2014)