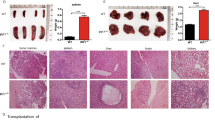
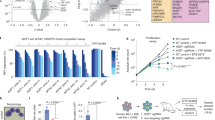
Abstract
Myeloid differentiation is blocked in acute myeloid leukemia (AML), but the molecular mechanisms are not well characterized. Meningioma 1 (MN1) is overexpressed in AML patients and confers resistance to all-trans retinoic acid-induced differentiation. To understand the role of MN1 as a transcriptional regulator in myeloid differentiation, we fused transcriptional activation (VP16) or repression (M33) domains with MN1 and characterized these cells in vivo. Transcriptional activation of MN1 target genes induced myeloproliferative disease with long latency and differentiation potential to mature neutrophils. A large proportion of differentially expressed genes between leukemic MN1 and differentiation-permissive MN1VP16 cells belonged to the immune response pathway like interferon-response factor (Irf) 8 and Ccl9. As MN1 is a cofactor of MEIS1 and retinoic acid receptor alpha (RARA), we compared chromatin occupancy between these genes. Immune response genes that were upregulated in MN1VP16 cells were co-targeted by MN1 and MEIS1, but not RARA, suggesting that myeloid differentiation is blocked through transcriptional repression of shared target genes of MN1 and MEIS1. Constitutive expression of Irf8 or its target gene Ccl9 identified these genes as potent inhibitors of murine and human leukemias in vivo. Our data show that MN1 prevents activation of the immune response pathway, and suggest restoration of IRF8 signaling as therapeutic target in AML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Ley TJ . Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 369: 98–98.
Rosenbauer F, Tenen DG . Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 2007; 7: 105–117.
Milligan DW, Wheatley K, Littlewood T, Craig JI, Burnett AK . Fludarabine and cytosine are less effective than standard ADE chemotherapy in high-risk acute myeloid leukemia, and addition of G-CSF and ATRA are not beneficial: results of the MRC AML-HR randomized trial. Blood 2006; 107: 4614–4622.
Lo-Coco F, Avvisati G, Vignetti M, Breccia M, Gallo E, Rambaldi A et al. Front-line treatment of acute promyelocytic leukemia with AIDA induction followed by risk-adapted consolidation for adults younger than 61 years: results of the AIDA-2000 trial of the GIMEMA Group. Blood 2010; 116: 3171–3179.
Heuser M, Argiropoulos B, Kuchenbauer F, Yung E, Piper J, Fung S et al. MN1 overexpression induces acute myeloid leukemia in mice and predicts ATRA resistance in patients with AML. Blood 2007; 110: 1639–1647.
Buijs A, Sherr S, van Baal S, van Bezouw S, van der Plas D, Geurts van Kessel A et al. Translocation (12;22) (p13;q11) in myeloproliferative disorders results in fusion of the ETS-like TEL gene on 12p13 to the MN1 gene on 22q11. Oncogene 1995; 10: 1511–1519.
Heuser M, Wingen LU, Steinemann D, Cario G, von Neuhoff N, Tauscher M et al. Gene-expression profiles and their association with drug resistance in adult acute myeloid leukemia. Haematologica 2005; 90: 1484–1492.
Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood 2004; 104: 3679–3687.
Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004; 350: 1617–1628.
Heuser M, Beutel G, Krauter J, Dohner K, von Neuhoff N, Schlegelberger B et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood 2006; 108: 3898–3905.
Carella C, Bonten J, Sirma S, Kranenburg TA, Terranova S, Klein-Geltink R et al. MN1 overexpression is an important step in the development of inv(16) AML. Leukemia 2007; 21: 1679–1690.
Haferlach C, Kern W, Schindela S, Kohlmann A, Alpermann T, Schnittger S et al. Gene expression of BAALC, CDKN1B, ERG, and MN1 adds independent prognostic information to cytogenetics and molecular mutations in adult acute myeloid leukemia. Genes Chromosomes Cancer 2012; 51: 257–265.
Langer C, Marcucci G, Holland KB, Radmacher MD, Maharry K, Paschka P et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol 2009; 27: 3198–3204.
Schwind S, Marcucci G, Kohlschmidt J, Radmacher MD, Mrozek K, Maharry K et al. Low expression of MN1 associates with better treatment response in older patients with de novo cytogenetically normal acute myeloid leukemia. Blood 2011; 118: 4188–4198.
Miller BG, Stamatoyannopoulos JA . Integrative meta-analysis of differential gene expression in acute myeloid leukemia. PLoS One 2010; 5: e9466.
Liu T, Jankovic D, Brault L, Ehret S, Baty F, Stavropoulou V et al. Functional characterization of high levels of meningioma 1 as collaborating oncogene in acute leukemia. Leukemia 2010; 24: 601–612.
Kandilci A, Grosveld GC . Reintroduction of CEBPA in MN1-overexpressing hematopoietic cells prevents their hyperproliferation and restores myeloid differentiation. Blood 2009; 114: 1596–1606.
Heuser M, Yun H, Berg T, Yung E, Argiropoulos B, Kuchenbauer F et al. Cell of origin in AML: susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer Cell 2011; 20: 39–52.
Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G . Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J 1998; 17: 3714–3725.
Slape C, Hartung H, Lin YW, Bies J, Wolff L, Aplan PD . Retroviral insertional mutagenesis identifies genes that collaborate with NUP98-HOXD13 during leukemic transformation. Cancer Res 2007; 67: 5148–5155.
Caudell D, Harper DP, Novak RL, Pierce RM, Slape C, Wolff L et al. Retroviral insertional mutagenesis identifies Zeb2 activation as a novel leukemogenic collaborating event in CALM-AF10 transgenic mice. Blood 2010; 115: 1194–1203.
Bergerson RJ, Collier LS, Sarver AL, Been RA, Lugthart S, Diers MD et al. An insertional mutagenesis screen identifies genes that cooperate with Mll-AF9 in a murine leukemogenesis model. Blood 2012; 119: 4512–4523.
Watanabe-Okochi N, Kitaura J, Ono R, Harada H, Harada Y, Komeno Y et al. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood 2008; 111: 4297–4308.
Braun CJ, Boztug K, Paruzynski A, Witzel M, Schwarzer A, Rothe M et al. Gene therapy for wiskott-Aldrich syndrome—long-term efficacy and genotoxicity. Sci Transl Med 2014; 6: 227ra233.
Li L, Jin H, Xu J, Shi Y, Wen Z . Irf8 regulates macrophage versus neutrophil fate during zebrafish primitive myelopoiesis. Blood 2010; 117: 1359–1369.
Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H et al. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood 2013; 121: 1839–1849.
Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K . ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity 2000; 13: 155–165.
Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 1996; 87: 307–317.
Hara T, Schwieger M, Kazama R, Okamoto S, Minehata K, Ziegler M et al. Acceleration of chronic myeloproliferation by enforced expression of Meis1 or Meis3 in Icsbp-deficient bone marrow cells. Oncogene 2008; 27: 3865–3869.
Gurevich RM, Rosten PM, Schwieger M, Stocking C, Humphries RK . Retroviral integration site analysis identifies ICSBP as a collaborating tumor suppressor gene in NUP98-TOP1-induced leukemia. Exp Hematol 2006; 34: 1192–1201.
Schmidt M, Hochhaus A, Nitsche A, Hehlmann R, Neubauer A . Expression of nuclear transcription factor interferon consensus sequence binding protein in chronic myeloid leukemia correlates with pretreatment risk features and cytogenetic response to interferon-alpha. Blood 2001; 97: 3648–3650.
Schmidt M, Nagel S, Proba J, Thiede C, Ritter M, Waring JF et al. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood 1998; 91: 22–29.
Niederle N, Kloke O, Wandl UB, Becher R, Moritz T, Opalka B . Long-term treatment of chronic myelogenous leukemia with different interferons: results from three studies. Leuk Lymphoma 1993; 9: 111–119.
Hao SX, Ren R . Expression of interferon consensus sequence binding protein (ICSBP) is downregulated in Bcr-Abl-induced murine chronic myelogenous leukemia-like disease, and forced coexpression of ICSBP inhibits Bcr-Abl-induced myeloproliferative disorder. Mol Cell Biol 2000; 20: 1149–1161.
Deng M, Daley GQ . Expression of interferon consensus sequence binding protein induces potent immunity against BCR/ABL-induced leukemia. Blood 2001; 97: 3491–3497.
Nardi V, Naveiras O, Azam M, Daley GQ . ICSBP-mediated immune protection against BCR-ABL-induced leukemia requires the CCL6 and CCL9 chemokines. Blood 2009; 113: 3813–3820.
Burchert A, Cai D, Hofbauer LC, Samuelsson MK, Slater EP, Duyster J et al. Interferon consensus sequence binding protein (ICSBP; IRF-8) antagonizes BCR/ABL and down-regulates bcl-2. Blood 2004; 103: 3480–3489.
Qian Z, Fernald AA, Godley LA, Larson RA, Le Beau MM . Expression profiling of CD34+ hematopoietic stem/ progenitor cells reveals distinct subtypes of therapy-related acute myeloid leukemia. Proc Natl Acad Sci USA 2002; 99: 14925–14930.
Otto N, Manukjan G, Gohring G, Hofmann W, Scherer R, Luna JC et al. ICSBP promoter methylation in myelodysplastic syndromes and acute myeloid leukaemia. Leukemia 2011; 25: 1202–1207.
Starczynowski DT, Karsan A . Deregulation of innate immune signaling in myelodysplastic syndromes is associated with deletion of chromosome arm 5q. Cell Cycle 2010; 9: 855–856.
Wei Y, Chen R, Dimicoli S, Bueso-Ramos C, Neuberg D, Pierce S et al. Global H3K4me3 genome mapping reveals alterations of innate immunity signaling and overexpression of JMJD3 in human myelodysplastic syndrome CD34+ cells. Leukemia 2013; 27: 2177–2186.
Schambach A, Wodrich H, Hildinger M, Bohne J, Krausslich HG, Baum C . Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol Ther 2000; 2: 435–445.
Heuser M, Yap DB, Leung M, de Algara TR, Tafech A, McKinney S et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood 2009; 113: 1432–1443.
Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q et al. H2 A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2012; 12: 180–192.
Ji X, Li W, Song J, Wei L, Liu XS . CEAS: cis-regulatory element annotation system. Nucleic Acids Res 2006; 34 (Web Server issue): W551–W554.
Krebs A, Frontini M, Tora L . GPAT: retrieval of genomic annotation from large genomic position datasets. BMC Bioinformatics 2008; 9: 533.
Huang da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
van Wely KH, Molijn AC, Buijs A, Meester-Smoor MA, Aarnoudse AJ, Hellemons A et al. The MN1 oncoprotein synergizes with coactivators RAC3 and p300 in RAR-RXR-mediated transcription. Oncogene 2003; 22: 699–709.
Wang ZY, Chen Z . Acute promyelocytic leukemia: from highly fatal to highly curable. Blood 2008; 111: 2505–2515.
Ficara F, Crisafulli L, Lin C, Iwasaki M, Smith KS, Zammataro L et al. Pbx1 restrains myeloid maturation while preserving lymphoid potential in hematopoietic progenitors. J Cell Sci 2013; 126 (Pt 14): 3181–3191.
Kocabas F, Zheng J, Thet S, Copeland NG, Jenkins NA, DeBerardinis RJ et al. Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood 2012; 120: 4963–4972.
Unnisa Z, Clark JP, Roychoudhury J, Thomas E, Tessarollo L, Copeland NG et al. Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood 2012; 120: 4973–4981.
Acknowledgements
We thank Dr AM Baru and PD Dr Michael Morgan for discussions and Renate Schottmann and Martin Wichmann for technical help. We also thank Dr Matthias Ballmaier and the staff of the Cell Sorting Core Facility of Hannover Medical School for their excellent service (supported in part by the Braukmann-Wittenberg-Herz-Stiftung). This study was supported by DFG grants HE 5240/5-1, grants DJCLS H09/1f, R 10/22, SP 12/02 and R13/14 from Deutsche-José-Carreras Leukämie-Stiftung e.V., grants 109003, 110284, 110292 and 111267 from Deutsche Krebshilfe and the German Federal Ministry of Education and Research grant 01EO0802 (IFB-Tx).
Author contributions
A Sharma and MH designed the research; A Sharma, HY, NJ, AC, A Schwarzer, EY, CKL, FK, BA, KG and MH performed the research; and A Sharma, HY, NJ, AC, A Schwarzer, EY, CKL, FK, BA, KG, AG, RKH and MH analyzed the data. A Sharma, HY and MH wrote the manuscript. All authors read and agreed to the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Sharma, A., Yun, H., Jyotsana, N. et al. Constitutive IRF8 expression inhibits AML by activation of repressed immune response signaling. Leukemia 29, 157–168 (2015). https://doi.org/10.1038/leu.2014.162
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/leu.2014.162
This article is cited by
-
ILC1s control leukemia stem cell fate and limit development of AML
Nature Immunology (2022)
-
LSD1 inhibition by tranylcypromine derivatives interferes with GFI1-mediated repression of PU.1 target genes and induces differentiation in AML
Leukemia (2019)
-
JMJD3 facilitates C/EBPβ-centered transcriptional program to exert oncorepressor activity in AML
Nature Communications (2018)
-
All-trans retinoic acid and arsenic trioxide fail to derepress the monocytic differentiation driver Irf8 in acute promyelocytic leukemia cells
Cell Death & Disease (2017)
-
Meis2 as a critical player in MN1-induced leukemia
Blood Cancer Journal (2017)