Abstract
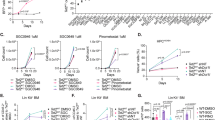
The inhibitor of apoptosis protein Survivin regulates hematopoiesis, although its mechanisms of regulation of hematopoietic stem cells (HSCs) remain largely unknown. While investigating conditional Survivin deletion in mice, we found that Survivin was highly expressed in phenotypically defined HSCs, and Survivin deletion in mice resulted in significantly reduced total marrow HSCs and hematopoietic progenitor cells. Transcriptional analysis of Survivin−/− HSCs revealed altered expression of multiple genes not previously linked to Survivin activity. In particular, Survivin deletion significantly reduced expression of the Evi-1 transcription factor indispensable for HSC function, and the downstream Evi-1 target genes Gata2, Pbx1 and Sall2. The loss of HSCs following Survivin deletion and impaired long-term HSC repopulating function could be partially rescued by ectopic Evi-1 expression in Survivin −/− HSCs. These data demonstrate that Survivin partially regulates HSC function by modulating the Evi-1 transcription factor and its downstream targets and identify new genetic pathways in HSCs regulated by Survivin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Altieri DC . Validating survivin as a cancer therapeutic target. Nat Rev Cancer 2003; 3: 46–54.
Altieri DC . Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer 2008; 8: 61–70.
Fukuda S, Pelus LM . Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther 2006; 5: 1087–1098.
Carter BZ, Milella M, Altieri DC, Andreeff M . Cytokine-regulated expression of survivin in myeloid leukemia. Blood 2001; 97: 2784–2790.
Adida C, Recher C, Raffoux E, Daniel MT, Taksin AL, Rousselot P et al. Expression and prognostic significance of survivin in de novo acute myeloid leukaemia. Br J Haematol 2000; 111: 196–203.
Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C et al. Analysis of human transcriptomes. Nat Genet 1999; 23: 387–388.
Fukuda S, Foster RG, Porter SB, Pelus LM . The antiapoptosis protein survivin is associated with cell cycle entry of normal cord blood CD34(+) cells and modulates cell cycle and proliferation of mouse hematopoietic progenitor cells. Blood 2002; 100: 2463–2471.
Fukuda S, Mantel CR, Pelus LM . Survivin regulates hematopoietic progenitor cell proliferation through p21WAF1/Cip1-dependent and -independent pathways. Blood 2004; 103: 120–127.
Xing Z, Conway EM, Kang C, Winoto A . Essential role of survivin, an inhibitor of apoptosis protein, in T cell development, maturation, and homeostasis. J Exp Med 2004; 199: 69–80.
Song J, So T, Cheng M, Tang X, Croft M . Sustained survivin expression from OX40 costimulatory signals drives T cell clonal expansion. Immunity 2005; 22: 621–631.
Okada H, Bakal C, Shahinian A, Elia A, Wakeham A, Suh WK et al. Survivin loss in thymocytes triggers p53-mediated growth arrest and p53-independent cell death. J Exp Med 2004; 199: 399–410.
Altznauer F, Martinelli S, Yousefi S, Thurig C, Schmid I, Conway EM et al. Inflammation-associated cell cycle-independent block of apoptosis by survivin in terminally differentiated neutrophils. J Exp Med 2004; 199: 1343–1354.
Gurbuxani S, Xu Y, Keerthivasan G, Wickrema A, Crispino JD . Differential requirements for survivin in hematopoietic cell development. Proc Natl Acad Sci USA 2005; 102: 11480–11485.
Blanc-Brude OP, Mesri M, Wall NR, Plescia J, Dohi T, Altieri DC . Therapeutic targeting of the survivin pathway in cancer: initiation of mitochondrial apoptosis and suppression of tumor-associated angiogenesis. Clin Cancer Res 2003; 9: 2683–2692.
Mesri M, Morales-Ruiz M, Ackermann EJ, Bennett CF, Pober JS, Sessa WC et al. Suppression of vascular endothelial growth factor-mediated endothelial cell protection by survivin targeting. Am J Pathol 2001; 158: 1757–1765.
Fukuda S, Singh P, Moh A, Abe M, Conway EM, Boswell HS et al. Survivin mediates aberrant hematopoietic progenitor cell proliferation and acute leukemia in mice induced by internal tandem duplication of Flt3. Blood 2009; 114: 394–403.
Leung CG, Xu Y, Mularski B, Liu H, Gurbuxani S, Crispino JD . Requirements for survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. J Exp Med 2007; 204: 1603–1611.
Pelus LM, Bian H, King AG, Fukuda S . Neutrophil-derived MMP-9 mediates synergistic mobilization of hematopoietic stem and progenitor cells by the combination of G-CSF and the chemokines GRObeta/CXCL2 and GRObetaT/CXCL2delta4. Blood 2004; 103: 110–119.
Buonamici S, Li D, Chi Y, Zhao R, Wang X, Brace L et al. EVI1 induces myelodysplastic syndrome in mice. J Clin Invest 2004; 114: 713–719.
Huang dW, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2007; 2: 2366–2382.
Fukuda S, Pelus LM . Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(+) cells by hematopoietic growth factors: implication of survivin expression in normal hematopoiesis. Blood 2001; 98: 2091–2100.
Fukuda S, Pelus LM . Elevation of Survivin levels by hematopoietic growth factors occurs in quiescent CD34+ hematopoietic stem and progenitor cells before cell cycle entry. Cell Cycle 2002; 1: 322–326.
Conway EM, Pollefeyt S, Cornelissen J, DeBaere I, Steiner-Mosonyi M, Ong K et al. Three differentially expressed survivin cDNA variants encode proteins with distinct antiapoptotic functions. Blood 2000; 95: 1435–1442.
Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci USA 2005; 102: 9194–9199.
Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR . A stem cell molecular signature. Science 2002; 298: 601–604.
Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994; 371: 221–226.
Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood 2005; 106: 477–484.
Shimabe M, Goyama S, Watanabe-Okochi N, Yoshimi A, Ichikawa M, Imai Y et al. Pbx1 is a downstream target of Evi-1 in hematopoietic stem/progenitors and leukemic cells. Oncogene 2009; 28: 4364–4374.
Ficara F, Murphy MJ, Lin M, Cleary ML . Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell 2008; 2: 484–496.
Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell 2008; 3: 207–220.
Chai L . The role of HSAL (SALL) genes in proliferation and differentiation in normal hematopoiesis and leukemogenesis. Transfusion 2011; 51 (Suppl 4): 87S–93S.
Berry FB, O'Neill MA, Coca-Prados M, Walter MA . FOXC1 transcriptional regulatory activity is impaired by PBX1 in a filamin A-mediated manner. Mol Cell Biol 2005; 25: 1415–1424.
Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat Genet 2005; 37: 225–232.
Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE . Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 2011; 333: 218–221.
Kostrouchova M, Kostrouch Z, Saudek V, Piatigorsky J, JE Rall . BIR-1, a Caenorhabditis elegans homologue of Survivin, regulates transcription and development. Proc Natl Acad Sci USA 2003; 100: 5240–5245.
Asanuma K, Tsuji N, Endoh T, Yagihashi A, Watanabe N . Survivin enhances Fas ligand expression via up-regulation of specificity protein 1-mediated gene transcription in colon cancer cells. J Immunol 2004; 172: 3922–3929.
Takizawa BT, Uchio EM, Cohen JJ, Wheeler MA, Weiss RM . Downregulation of survivin is associated with reductions in TNF receptors’ mRNA and protein and alterations in nuclear factor kappa B signaling in urothelial cancer cells. Cancer Invest 2007; 25: 678–684.
Balkhi MY, Christopeit M, Chen Y, Geletu M, Behre G . AML1/ETO-induced survivin expression inhibits transcriptional regulation of myeloid differentiation. Exp Hematol 2008; 36: 1449–1460.
Kataoka K, Sato T, Yoshimi A, Goyama S, Tsuruta T, Kobayashi H et al. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J Exp Med 2011; 208: 2403–2416.
Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL et al. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol 2000; 10: 1319–1328.
Yoshimi A, Goyama S, Watanabe-Okochi N, Yoshiki Y, Nannya Y, Nitta E et al. Evi1 represses PTEN expression and activates PI3K/AKT/mTOR via interactions with polycomb proteins. Blood 2011; 117: 3617–3628.
Carter BZ, Qiu Y, Huang X, Diao L, Zhang N, Coombes KR et al. Survivin is highly expressed in CD34(+)38(−) leukemic stem/progenitor cells and predicts poor clinical outcomes in AML. Blood 2012; 120: 173–180.
Persons DA, Allay JA, Allay ER, Ashmun RA, Orlic D, Jane SM et al. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood 1999; 93: 488–499.
Altieri DC . Survivin and IAP proteins in cell-death mechanisms. Biochem J 2010; 430: 199–205.
Salz W, Eisenberg D, Plescia J, Garlick DS, Weiss RM, Wu XR et al. A survivin gene signature predicts aggressive tumor behavior. Cancer Res 2005; 65: 3531–3534.
Broxmeyer HE, Lu L, Platzer E, Feit C, Juliano L, Rubin BY . Comparative analysis of the influences of human gamma, alpha and beta interferons on human multipotential (CFU-GEMM), erythroid (BFU-E) and granulocyte-macrophage (CFU-GM) progenitor cells. J Immunol 1983; 131: 1300–1305.
Pelus LM, Ottmann OG, Nocka KH . Synergistic inhibition of human marrow granulocyte-macrophage progenitor cells by prostaglandin E and recombinant interferon-alpha, -beta, and -gamma and an effect mediated by tumor necrosis factor. J Immunol 1988; 140: 479–484.
Magee WE, Griffith MJ . The liver as a site for interferon production in response to poly I:poly C. Life Sci II 1972; 11: 1081–1086.
Manetti R, Annunziato F, Tomasevic L, Gianno V, Parronchi P, Romagnani S et al. Polyinosinic acid: polycytidylic acid promotes T helper type 1-specific immune responses by stimulating macrophage production of interferon-alpha and interleukin-12. Eur J Immunol 1995; 25: 2656–2660.
Acknowledgements
We thank Susan Rice and Denessa Lucket for cell sorting. This work was supported by a Biomedical Research Grant from the Indiana University School of Medicine, Research Support Funds Grants from Indiana University and Purdue University, Indianapolis, Research Support Funds from Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Sankyo Biomedical Research Foundation and Mitsubishi Pharma Research Foundation, the Japan Leukaemia Research Fund, an AstraZeneca Research Grant, the Naito Memorial Foundation, a Grant-in-Aid for Scientific Research (B) (20390298) from the Japan Society for the Promotion of Science (to SF) and US Public Health Service Grants (HL69669, HL079654 and HL096305) from the National Institutes of Health (to LMP). EMC is an adjunct scientist with the Canadian Blood Services and holds a Canada Research Chair in Endothelial Biology and a CSL-Behring Research Chair.
Author Contributions
SF directed the projects, designed the research, performed experiments, analyzed the data and wrote the manuscript; JH performed experiments, analyzed data and wrote the manuscript; PS, JMS, MA and SY performed experiments and analyzed the data; EMC and GN prepared materials; LMP directed the project, performed experiments, interpreted the data and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Fukuda, S., Hoggatt, J., Singh, P. et al. Survivin modulates genes with divergent molecular functions and regulates proliferation of hematopoietic stem cells through Evi-1. Leukemia 29, 433–440 (2015). https://doi.org/10.1038/leu.2014.183
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/leu.2014.183
This article is cited by
-
T-cell-based immunotherapy of acute myeloid leukemia: current concepts and future developments
Leukemia (2021)
-
Hematopoietic Stem Cell Mobilization: Current Collection Approaches, Stem Cell Heterogeneity, and a Proposed New Method for Stem Cell Transplant Conditioning
Stem Cell Reviews and Reports (2021)
-
Emerging Importance of Survivin in Stem Cells and Cancer: the Development of New Cancer Therapeutics
Stem Cell Reviews and Reports (2020)
-
Inhibitors of apoptosis: clinical implications in cancer
Apoptosis (2017)
-
Survivin: a unique target for tumor therapy
Cancer Cell International (2016)