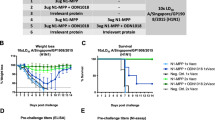
Abstract
Influenza A remains a significant public health challenge because of the emergence of antigenically shifted or highly virulent strains1,2,3,4,5. Antiviral resistance to available drugs such as adamantanes or neuraminidase inhibitors has appeared rapidly6,7,8,9, creating a need for new antiviral targets and new drugs for influenza virus infections. Using forward chemical genetics, we have identified influenza A nucleoprotein (NP) as a druggable target and found a small-molecule compound, nucleozin, that triggers the aggregation of NP and inhibits its nuclear accumulation. Nucleozin impeded influenza A virus replication in vitro with a nanomolar median effective concentration (EC50) and protected mice challenged with lethal doses of avian influenza A H5N1. Our results demonstrate that viral NP is a valid target for the development of small-molecule therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Webster, R.G. & Govorkova, E.A. H5N1 influenza – continuing evolution and spread. N. Engl. J. Med. 355, 2174–2177 (2006).
Regoes, R.R. & Bonhoeffer, S. Emergence of drug-resistant influenza virus: population dynamical considerations. Science 312, 389–391 (2006).
Moscona, A. Global transmission of oseltamivir-resistant influenza. N. Engl. J. Med. 360, 953–956 (2009).
Dharan, N.J. et al. Oseltamivir-Resistance Working Group. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. J. Am. Med. Assoc. 301, 1034–1041 (2009).
Layne, S.P., Monto, A.S. & Taubenberger, J.K. Pandemic influenza: an inconvenient mutation. Science 323, 1560–1561 (2009).
Lackenby, A., Thompson, C.I. & Democratis, J. The potential impact of neuraminidase inhibitor resistant influenza. Curr. Opin. Infect. Dis. 21, 626–638 (2008).
Yuen, K.Y. et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351, 467–471 (1998).
Cumulative Number of Confirmed Human Cases of Avian Influenza A (H5N1) Reported to WHO (http://www.who.int/csr/disease/avian_influenza/country/cases_table_2009_09_24/en/index.html).
Wong, S.S. & Yuen, K.Y. Avian influenza virus infections in humans. Chest 129, 156–168 (2006).
Kao, R.Y. et al. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 11, 1293–1299 (2004).
Kao, R.Y. et al. Characterization of SARS-CoV main protease and identification of biologically active small molecule inhibitors using a continuous fluorescence-based assay. FEBS Lett. 576, 325–330 (2004).
Portela, A. & Digard, P. The influenza virus nucleoprotein: a multifunctional, RNA-binding protein pivotal to virus replication. J. Gen. Virol. 83, 723–734 (2002).
Davey, J., Dimmock, N.J. & Colman, A. Identification of the Sequence Responsible for the Nuclear Accumulation of the Influenza Virus. Cell 40, 667–675 (1985).
Boulo, S., Akarsu, H., Ruigrok, R.W.H. & Baudin, F. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 124, 12–21 (2007).
Ozawa, M. et al. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 81, 30–41 (2007).
Ye, Q., Krug, R.M. & Tao, Y.J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444, 1078–1082 (2006).
Hoffmann, E., Neumann, G., Kawaoka, Y., Hobom, G. & Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97, 6108–6113 (2000).
Wang, P. et al. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J. Virol. 83, 7850–7861 (2009).
Wu, W.W., Sun, Y.H. & Panté, N. Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol. J. 4, 49 (2007).
Digard, P. et al. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J. Virol. 73, 2222–2231 (1999).
Elton, D., Medcalf, E., Bishop, K., Harrison, D. & Digard, P. Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J. Virol. 73, 7357–7367 (1999).
Zheng, B.J. et al. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc. Natl. Acad. Sci. USA 105, 8091–8096 (2008).
Stockwell, B.R. Chemical genetics: ligand-based discovery of gene function. Nat. Rev. Genet. 1, 116–125 (2000).
Strausberg, R.L. & Schreiber, S.L. From knowing to controlling: a path from genomics to drugs using small molecule probes. Science 300, 294–295 (2003).
Min, J.Y. & Krug, R.M. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 103, 7100–7105 (2006).
Campanacci, V. et al. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc. Natl. Acad. Sci. USA 100, 5069–5074 (2003).
Morris, G.M. et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 (1998).
Brooks, R. et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983).
Gubareva, L.V. et al. Characterization of mutants of influenza A virus selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J. Virol. 70, 1818–1827 (1996).
Niepmann, M. & Zheng, J. Discontinuous native protein gel electrophoresis. Electrophoresis 27, 3949–3951 (2006).
Acknowledgements
This study was supported in part by the Carol Yu Center for Infection Seed Fund for Basic Research from the University of Hong Kong, the Research Fund for the Control of Infectious Diseases and the Area of Excellence Scheme of the University Grant Council (Grant AoE/M-12/06). The Beckman Coulter Core system is a generous gift from the Hong Kong Sanatorium Hospital Doctors' Donation Fund by Y.-C. Tsao, C.-M. Chan, G. Lo, K.-M. Lai, R.K.Y. Lo, M. Tsao, B.S.S. Tse, T.-F. Tse, S.W.C. Wu, D.Y.C. Yu, R.Y.H. Yu and Y.-K. Tsao. We are grateful to R. Webster for gifts of the pHW2000 plasmids and E. Hoffmann for luciferase reporter system. We thank V. Poon, C. Chan and Q. Zhang for mice studies and K.H. Chan for virus strains. The use of Confocal Systems Core Facility provided by the LKS Faculty of Medicine, HKU, is acknowledged.
Author information
Authors and Affiliations
Contributions
R.Y.K. and K.-Y.Y conceived the study. R.Y.K. designed and performed experiments and analyzed data. D.Y. gave conceptual advice and technical support on chemistry. L.-S.L., W.H.W.T., J.D., M.-P.C., C.-M.C. and P.W. performed experiments. J.S., L.H., and G.C. performed molecular dockings. B.-J.Z. provided animal study data. J.-D.H. gave conceptual advice on protein trafficking. J.M. constructed database and performed HTS data normalization. H.C. and Y.G. provided reverse genetics system. K.-Y.Y. did troubleshooting and provided the grant support. R.Y.K. and K.-Y.Y. supervised the study and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3 and Supplementary Figs. 1–11 (PDF 657 kb)
Rights and permissions
About this article
Cite this article
Kao, R., Yang, D., Lau, LS. et al. Identification of influenza A nucleoprotein as an antiviral target. Nat Biotechnol 28, 600–605 (2010). https://doi.org/10.1038/nbt.1638
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/nbt.1638
This article is cited by
-
Exploration of phytochemical compounds against Marburg virus using QSAR, molecular dynamics, and free energy landscape
Molecular Diversity (2023)
-
An anti-influenza A virus microbial metabolite acts by degrading viral endonuclease PA
Nature Communications (2022)
-
Development of a target identification approach using native mass spectrometry
Scientific Reports (2021)
-
Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism
Nature Microbiology (2020)
-
SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target
Nature Communications (2019)